Aside from transcription, shRNA maturation and activation is directly dependent on a number of proteins for its function. In cleavage of the initially transcribed shRNA into export-ready pre-shRNA, Drosha (RNAse III family member) is required. In its further cleavage in the cytosol, the RNAse III Dicer is required. Additionally, the protein TRBP/PACT heterodimer is required. Furthermore, the RISC protein Argonaute2 is the only enzyme known to unwind the dsRNA to render it competent to interact with its target messenger RNA.
As mentioned above, the nuclear export of shRNA likely occurs via the mechanism of miRNA, whereby Ran-GTP bound Exportin-5 is able to drive pre-shRNA into the cytosol. shRNA release is triggered by the hydrolysis of GTP to GDP, generating a unidirectional flow of pre-shRNAs into the cytosol (for a review of basic mechanism, see Rao et al. 2009).
shRNA can act via a miRNA- or siRNA-like mechanism. Either case drives the repression of translation, but it is believed that the miRNA-like mechanism is more rapid, albeit somewhat non-specific, while the siRNA-like mechanism is slower and acts via perfect complementarity for a target message (Rao et al. 2009), as discussed above. In both cases, it is the complementarity of the unwound, Argonaute-bound shRNA that causes binding of mRNA. This RNA hybridization drives the degradation of the message in the siRNA-like case, or preventation of translation in the miRNA-like mechanism. One proposed idea for the fate of such bound mRNA is that it is enzymatically cleaved, while another notion is that it is sequestered to the so-called p-body in the cytoplasm.
shRNA is a ribonucleic acid polymer that is designed based on the concepts garnered from the study of naturally-occurring hairpin RNAs involved in RNAi (namely, siRNA and miRNA). Its function in the cell is to drive the degradation of mRNAs in a sequence-specific manner. The recent intensive study of these molecules, however, implicates a number of direct and indirect side-effects of shRNA, including off-target repression of mRNAs, induction of an interferon response (Bauer et al. 2009), and competition with native RNAs for access to RNAi processing machinery (Grimm et al. 2006).
Bernards, R., T.R. Brummelkamp, R.L. Beijersbergen. (2006) shRNA libraries and their use in cancer genetics. Nature Methods. 3(9):701-6. [Pubmed]
Silva, J.M., M.Z. Li, K. Chang, W. Ge, M.C. Golding, R.J. Rickles, D. Siolas, G. Hu, P.J. Paddison, M.R. Schlabach, N. Sheth, J. Bradshaw, J. Burchard, A. Kulkarni, G. Cavet, R. Sachidanandam, W.R. McCombie, M.A. Cleary, S.J. Elledge, G.J. Hannon. (2005) Second-generation shRNA libraries covering the mouse and human genomes. Nature Genetics. (11): 1281-8. [Pubmed]
Yi, R., Y. Qin, I.G. Macara, B.R. Cullen. (2003) Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes and Development. 17(24): 3011-6. [Pubmed]
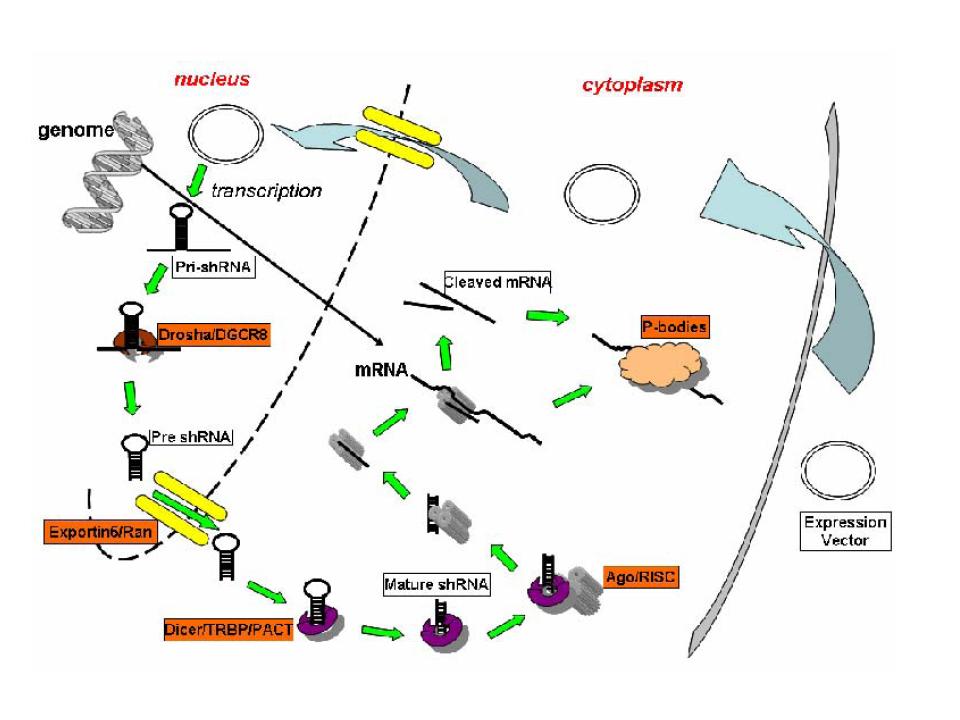
shRNAs are genetically engineered analogs of miRNAs. The way in which they are processed is believed to proceed as that of an miRNA, as outlined below. In essence, a long precursor RNA is catalytically cleaved by an RNAse III in the nucleus, transported to the cytoplasm in a GTP-dependent manner, cleaved again by an RNAse III, loaded onto an adaptor protein that unwinds the now short dsRNA to expose a single strand, and targeted to a cognate or near-cognate mRNA. Much of this process is poorly understood. The shRNA can be delivered "naked" (nucleic acid alone) or via a viral vector.
Specifically, shRNA biogenesis occurs initially via the delivery of the viral vector to the nucleus. Upon vector integration, the product is transcribed from either RNA pol II (e.g. T7) or pol III promoter (e.g. U6). The long, dsRNA, termed pre-shRNA, is then cleaved to a roughly 65 nucleotide (nt) fragment that consists of a 2 nt 3' overhang along with the characteristic hairpin loop structure and stem of complementary RNA base pairing. This initial cleavage is catalyzed by Drosha, an RNAse III. This process also appears to require DGCR8, a dsRNA binding protein. The hairpin is cleaved from the stem following nuclear export in a Ran-GTPase dependent manner. A Ran GTPase requirement for nuclear export has been noted for a number of other non-coding RNAs, such as tRNA. In the case of miRNA miR-30, the protein required for export is termed Exportin-5. The hydrolysis of GTP bound to Ran in the cytoplasm triggers the dissociation of the complex, allowing the pre-shRNA to bind Dicer, an RNAse III. Here the RNA undergoes further cleavage to the familiar ~22 nt length. These fragments with limited complementarity to the mRNA they target are then transferred to the Argonaute-containing RNA induced silencing complex (RISC), and Argonaute2 drives the unwinding of the dsRNA to yield a primed RISC with a guide sequence that is able to target specifically the region of an mRNA to which its shRNA fragment is complementary (for comparison to the biogenesis of siRNA, see Rao et al. 2009).
Permission received for reprint. (Rao et al. 2009)
Because there is theoretically no limitation to the number of mRNAs the researcher can target with shRNA, it's potential as a research tool is staggering. The large-scale screening of cell lines for novel players in common pathways has brought mammalian genetics to the level of more tractable, traditional cell-based genetics models (e.g. Berns et al. 2004). The shortcomings of RNAi in mammalian systems has largely been circumvented with the advent of genetically-encoded shRNA-expressing viral vectors that transmit through all cell generations within an organism's lifetime. Major issues with in vivo toxicity of shRNA has to be kept in mind during the experimental design, and controls that are sensitive to off-target, non-specific effects are an absolute must (see above). The ability to deliver shRNA as an in vivo means of generating hypomorphic alleles is likely to prove important in the advancement of our understanding of genetic loci that have proven difficult to modify with traditional genetic manipulations, e.g. homologous recombination (for discussion, see Dykxhoorn and Lieberman 2005). Additionally, the dose-specific arguments that are posited in many published works can now be bolstered by a careful shRNA "knockdown" approach in which a dose-dependency of a particular phenomenon can acutally be observed, rather than speculated upon.
In short, the greatest role for shRNA is ascertaining novel mammalian cell pathway components. The near future will likely bring a larger-scale, quantitative approach to understanding the protein interactome of normal and malignant mammalian cell lines, as has been seen with recently published yeast physical interaction maps. The ability to screen libraries of shRNA for a desired phenotypic effect is generating new data in fields that were thought to have been thoroughly mined.
Its limitations as a tool in fighting human disease are discussed below.
short hairpin RNA (shRNA) is an artificial form of RNA interference modeled after endogenous pathways.
RNA interference (RNAi) has a short history but has generated a windfall of paradigm-shifting scientific findings and numerous promising clinical products. 1990 saw the first publication of an RNAi mechanism, whereby Napoli, Lemieux, and Jorgensen noted the loss of expression of a pigmentation gene in Petunias following the attempted overexpression of that gene (Napoli et al. 1990). It was later in the decade that the first animal RNAi was discovered, again somewhat inadvertantly (Fire et al. 1998). This discovery engendered a revolution in the way scientists approach questions regarding animal gene regulation, and began to elucidate a very deeply conserved mechanism to regulate gene expression. The first papers outlining the utility of short hairpin RNAs came in 2002 (Paddison et al. [2002] and Brummelkamp et al. [2002]). These works marked the beginning of the artificial RNAi efforts that have led to very large scale genetic screens in mammalian cell culture (Dykxhoorn and Lieberman [2005]). These efforts have borne numerous fruit, such as the elucidation of a more complete roster of factors involved in the p53-mediated cell cycle arrest process (Berns et al. 2004). The rise of genomically-encoded shRNAs has allowed researchers to "knockdown" gene function in specific mouse tissues. More recently, shRNA has become a promising alternative to traditional disease fighting, with a number of si- and shRNA-therapies in clinical trials (Rao et al. 2009). The discovery that shRNAs can be efficiently delivered with viral vectors to cells (Rubinson et al. 2003) and in vivo (e.g. Giering et al. 2008) is predicted to help the rise of this technology as an important therapy (see Blow [2007] and Castanotto and Rossi [2009]).
shRNA is an important tool in the assessment of gene function in mammals, and is used largely as a research tool. Its applicability in human disease prevention and treatment is not well-understood, largely because of the difficulty of delivery of shRNA to the cells of the host. The alternative to delivering shRNA, as discussed above, is to provide the cell with short dsRNA, analogous to siRNA, that will be immediately taken up by RISC and targeted to mRNA. There are a number of clinical trials underway testing the efficacy of such modalities, with tissue targeting posing the biggest hurdle. An airborne delivery system used to treat certain lung conditions with non-viral siRNA has been successful in the clinic, but internal organs that are more difficult to access are proving a challenge.
A rare example of a clinical study using shRNA as a therapeutic is that of the joint Benitec/City of Hope Medical Center AIDS lymphoma case. A lentiviral vector expressing antagonists of two exons of the HIV genome has been integrated into the blood stem cells of 4 patients. The expectation is that the healthy, shRNA-transcribing cells will renew the blood cell population while knocking down the expression of 2 required HIV genes, tat and rec (Castanotto and Rossi 2009).

