siRNAs are thought to have most likely evolved as a primitive immune response evoked by the presence of foreign nucleic acids.[6] siRNA or siRNA-like molecules (such as rasiRNA) are represented in nearly every kingdom (the exception being bacteria), including algae (Chlamydomonas reinhardtii), plants (A. thaliana), fungi (S. pombe) and both vertebrate and invertebrate members of the animal kingdom (D. melanogaster, C. elegans, M. musculus, H. sapiens). [7] Whereas humans only express one isoform of the Dicer enzyme, Drosophila and Arabidopsis express two and four Dicer paralogues (known as Dicer-like proteins or DCLs) respectively. Knockout studies in these species support the notion that there is functional redundancy amongst these DCL paralogues. The same may be true of the RISC component Argonaute; Ago2 is the sole member of this protein family expressed in higher mammals, while lower eukaryotes seems to make use of multiple members.[7]
See Biogenesis
Besides its useful ability to silence genes via translational inhibition, the primary cellular functions of siRNA are protection from exogenous (foreign) nucleic acids (e.g. viruses), and the maintenance of genome integrity via transcriptional silencing of undesired genomic loci (e.g. retrotransposons or repeat sequences). [9]
In Arabidopsis thaliana, siRNA-mediated cytosine methylation has been shown to play an active role in chromatin remodeling and to be crucial for the maintenance of the heterochromatic state. [12]
In the protozoan Tetrahymena thermophila, the siRNA family member scnRNA is involved in DNA elimination of genomic loci in the micronuclei during conjugation and macronuclear formation. [16]
- Dykxhoorn D, Novina C, Sharp P (2003). "Killing the messenger: short RNAs that silence gene expression". Nature Reviews Molecular Cell Biology 4(6): 457-67. PMID: 12778125
- Dorsett Y, Tuschl T (2004). "siRNAs: Applications in functional genomics and potential as therapeutics". Nature Reviews Drug Discovery 3(4): 318-329. doi:10.1038/nrd1345 PMID: 15060527 .
- Gannon, G (2002). "RNA interference". Nature 418: 244-251. PMID: 12110901.
1. Chalk A, Wahlestedt C., Sonnhammer E (2004). "Improved and automated prediction of effective siRNA". Biochemical and Biophysical Research Communications 319(1): 264-274. ![]() doi:10.1016/j.bbrc.2004.04.181 PMID: 17415516
doi:10.1016/j.bbrc.2004.04.181 PMID: 17415516
2. Fire A, Xu S, Montgomery M, Kostas S, Driver S, Mello C (1998). "Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans". Nature 391 (6669): 806–11. doi:. PMID 9486653.
3. Hamilton A, Baulcombe D (1999). "A species of small antisense RNA in posttranscriptional gene silencing in plants". Science 286 (5441): 950–2. doi:. PMID 10542148.
4. Elbashir S, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T (2001). "Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells". Nature 411 (6836): 494–8. doi:. PMID 11373684.
5. Naqvi A, Islam N, Choudhury N, Haq, Q (2009). "The fascinating world of RNA interference". International Journal of Biological Sciences 5(2): 97-117. http://www.biolsci.org/v05p0097.htm PMID: 19173032.
6. Marques J, Williams B (2005). "Activation of the mammalian immune system by siRNAs". Nature Biotechnology 23: 1399-1405. PMID: 16273073.
7. Meister G, Tuschl T (2004). "Mechanisms of gene silencing by double-stranded RNA". Nature 431: 343-349. PMID: 15372041
8. Hannon G, Rossi J (2004). "Unlocking the potential of the human genome with RNA interference". Nature 431 (7006): 371–8. doi:. PMID 15372045. http://www.nature.com/nature/journal/v431/n7006/full/nature02870.html.
9. Elbashir S, Lendeckel W, Tuschl T (2001). "RNA Interference is mediated by 21- and 22- nucleotide RNAs". Genes Dev 15 (2): 188–200. doi:. PMID 11157775. http://genesdev.cshlp.org/cgi/content/full/15/2/188.
10. Shi Y (2002). "Mammalian RNAi for the masses". Trends in Genetics 19 (1): 9-12. ![]() doi:10.1016/S0168-9525(02)00005-7 PMID: 12493242.
doi:10.1016/S0168-9525(02)00005-7 PMID: 12493242.
11. Xia H, Mao Q, Paulson H, Davidson B (2002). "siRNA-mediated gene silencing in vitro and in vivo". Nature Biotechnology 20: 1006-1010. PMID: 12244328.
12. Pontes O, Li C, Nunes P, Haag J, Ream T, Vitins A, Jacobsen S, Pikaard C (2008). "The Arabidopsis chromatin-modifying nuclear siRNA pathway involves a nucelolar RNA processing center". Cell 126(1): 79-92. PMID: 16839878.
13. Akashi H, Matsumoto S, Taira K (2005). "Gene discovery by ribozyme and siRNA libraries". Nature Reviews Molecular Cell Biology 6: 413-422. PMID: 15956980.
14. Katiyar-Agarwal S, Morgan R, Dahlbeck D, Borsani O, Villegas A, Zhu J, Staskawicz B, Jin H (2006). "A pathogen-inducible endogenous siRNA in plant immunity". PNAS 103(47): 18002-8007. PMCID: PMC1693862.
15. Miller T, Kaspar B, Kops G, Yamanaka K, Christian L, Gage F, Cleveland D (2006). "Virus delivered small RNA silencing sustains strength in amyotrophic lateral sclerosis". Annals of Neurology 57: 773-776. PMID: 15852369
16. Liu Y, Mochizuki K, Gorovsky M (2004). "Histone H3 lysine 9 methylation is required for DNA elimination in developing macronuclei in Tetrahymena". PNAS 101(6): 1679-84. PMID: 14755052
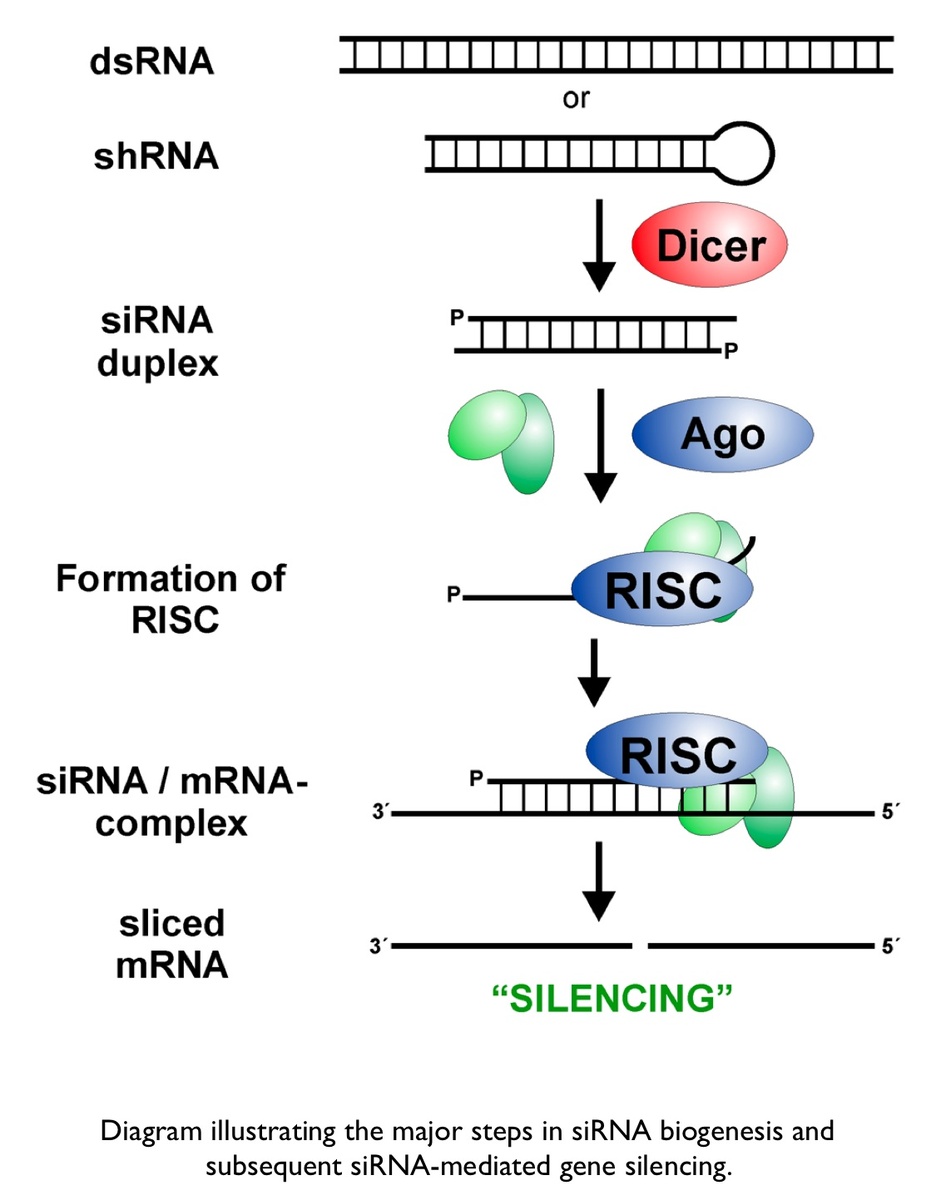
The cascade leading to the generation of mature siRNA begins with transcription by RNA polymerase II (in animals), RNA polymerase III (from a shRNA template), or RNA polymerase IV (in plants), forming double stranded RNA (dsRNA). These dsRNA are recognized by the RNA binding domain (RBD) of the Dicer complex, which contains a pair of RNaseIII type endonuclease catalytic domains. The dimeric Dicer complex then cleaves the dsRNA into ~21-28 nucleotide siRNA duplexes containing 2-nucleotide 3' overhangs with 5' phosphate and 3' hydroxyl termini, which are bound by the Dicer PAZ (Piwi Argonaute Zwille) domain.
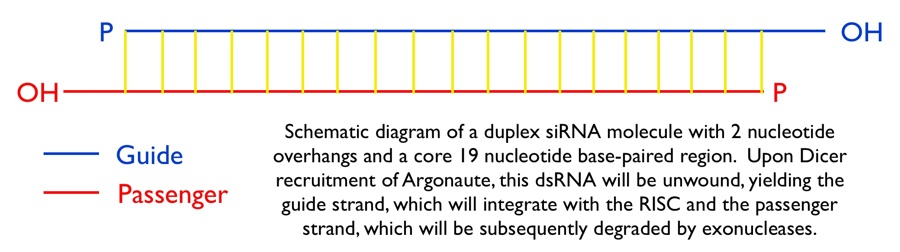
Subtle species-specific variations in the spacing of the Dicer catalytic domains have been proposed as an explanation as to why Dicer-cleaved RNA species range from 19-28 nucleotides in length.[8] After processing at the hands of Dicer, the siRNA-Dicer complex is exported through the nuclear pore complex (NPC) into the cytosol, where Dicer recruits the protein Argonaute and the RISC is assembled. Argonaute has helicase activity and the ability to detect the thermal stability of duplex siRNA ends. Argonaute identifies the end with lower thermal stability and proceeds to unwind the duplex siRNA, generating two single-stranded siRNAs. The strand that is retained as a functional component of the RNA Induced Silencing Complex (RISC) is referred to as the guide strand, whereas the other strand, which will be rapidly degraded by exonucleases, is known as the passenger strand. The guide strand recruits the RISC to the surface of mRNAs that are homologous to the siRNA sequence. siRNA recognition of the target mRNA is conferred by the "seed region", a six nucleotide stretch corresponding to positions 2-7 on the antisense siRNA strand. After the siRNA seed region anneals, the catalytic RNase H domain of Argonaute then subjects perfectly complementary mRNA sequences 10 nucleotides from the 5' end of the incorporated siRNA strand to nucleolytic degradation, resulting in the translational inhibition of the target mRNA.
Some members of the siRNA family include trans-acting small interfering RNAs (tasiRNAs), repeat-associated small interfering RNAs (rasiRNAs), scan RNAs (scnRNAs) and long siRNAs (lsiRNAs). Micro RNAs (miRNAs), piwi-interacting RNAs (piRNAs) and 21U-RNAs are also debatably members of the siRNA class, albeit with different biogenesis pathways and/or downstream effects.[5]
siRNAs hold nearly limitless potential for use as a tool, which has led to their rise as the most widely used gene-silencing approach. To date, their use has been cited in thousands of publications. The most popular utilizations of siRNA are to synthetically produce siRNAs and introduce them into cells via injection or electroporation or to express them from vectors that transcribe them as short hairpin RNAs (shRNAs) that are processed by the cell's internal machinery into siRNAs. [10] Vector-based siRNA delivery allows for long-term knockdown of target transcripts since the vector is able to stably integrate into the genome. In contrast, synthetically produced injected/transfected siRNAs yield a more transient knockdown of gene activity that is largely dependent on the rate of cell division. siRNAs have also been incorporated into adenoviral, adeno-associated viral, retroviral and lentiviral vectors. [11] The viral-based siRNA expression system holds great promise as a form of gene therapy for human diseases and as a more general tool to study gene function in vivo. One of the great advantages of siRNA as a genetic tool is the ability to study functional hypomorphic alleles that would be lethal to the organism if completely abrogated with knockout technology. siRNA approaches have been widely used to elucidate the roles of unknown genes, link genes into causal pathways, and in the case of genome-wide siRNA screens, to discover novel genes. [13]
Small interfering RNA (siRNA) is also known as short interfering RNA or silencing RNA. In the literature, synthetic siRNA constructs are generally denoted by "gene name" - siRNA (e.g. p53-siRNA), with constructs targeting the same gene distinguished by the addition of numbers after the construct name (e.g. p53-siRNA-1 and p53-siRNA-2). Base numbering in siRNAs is referred to by the sense strand, with the first 5' nucleotide on the sense strand corresponding to the 19 position on the antisense strand.[1]

Andrew Fire, 2006 Nobel Laureate (Physiology/Medicine)
Although the broader phenomenon of RNA interference (RNAi) was first elucidated by Andrew Fire and Craig C. Mello in C. elegans in 1998,[2] discovery of siRNA as a specific endogenous effector molecule capable of RNA interference is generally credited to David Baulcombe and Andrew Hamilton, who reported siRNA as a novel small antisense RNA underlying a posttrascriptional gene silencing (PTSG) mechanism in plants.[3] Another major milestone, which highlighted the potential applications of siRNA, was the 2001 paper by Thomas Tuschl and colleagues demonstrating the capability of synthetic siRNAs to mediate RNA interference in mammalian cells.[4] This achievement led to the widespread use of synthetic siRNAs as a laboratory tool to selectively knock-down the activity of specific genes.
While it has not been definitively demonstrated that any endogenous siRNAs play a causal role in human disease, this does not necessarily preclude the existence of disease-mediating siRNAs. Indeed, an endogenous siRNA induced by the presence of the pathogen Pseudomonas syringae in plants indicates that there may be a conserved role for siRNAs in immune responses.[14] For the moment though, the broader implications of endogenous siRNAs in human disease remain unclear with the focus instead being on using synthetic siRNA approaches as a tool to elucidate genetic mechanisms and pathways relevant to disease. siRNA-based therapeutics are currently being actively pursued by many pharmaceutical and biotechnology companies in research areas including cancer, immunology and neurological disease. The legitimacy of siRNA therapeutic approaches was underscored by a recent study reporting that siRNA viral vectors may have promise in the treatment of amyotrophic lateral sclerosis (ALS).[15]

