Diversity and specificity in taxonomy and molecular mechanisms suggest that riboswitches may be one of the oldest of genetic control systems [12]. Of the two riboswitch domains, the aptamer is highly conserved in even distantly related organisms, while sequence and structure are far more varied in the expression platform. Like protein receptors, riboswitch aptamers are highly selective and specific, indicating that the performance characteristics of riboswitches are competitive with those that are exhibited by proteins. Their ability to specifically bind small molecules and catalyze reactions lends support to the RNA world hypothesis, which suggests that an ancient world based on RNA existed before DNA-based organisms [5].
Labs that study riboswitches:
Bioinformatic Tools to study riboswitches:
mfold: software for folding nucleic acids
- TPP riboswitch binds thiamine pyrophosphate to regulate thiamin biosynthesis and transport [13].
- FMN riboswitches bind flavin mononucleotide to regulate riboflavin biosynthesis and transport [14].
- Cobalamin riboswitches bind adenosylcobalamin (the coenzyme form of vitamin B12) to regulate cobalamin biosynthesis and transport [16].
- SAM riboswitches bind S-adenosylmethionine to regulate methionine and SAM biosynthesis and transport [6, R2].
- Three distinct SAM riboswitches are known: SAM-I, SAM-II and the SMK riboswitch.
- A related riboswitch, the SAH riboswitch binds S-adenosylhomocysteine to regulate genes involved in recycling this metabolite, produced when S-adenosylmethionine is used in methylation reactions.
- preQ1 riboswitches bind pre-quenosine-1 to regulate genes involved in the synthesis or transport of this precursor to quenosine [17, R2].
- Two entirely distinct classes of preQ1 riboswitches are known: preQ1-I riboswitches and preQ1-II riboswitches. The binding domain of preQ1-I riboswitches are unusually small among naturally occurring riboswitches, while preQ1-II riboswitches have a completely different structure and are larger.
- Purine riboswitches bind either guanine or adenine to regulate purine metabolism and transport [18].
- Lysine riboswitches bind lysine to regulate lysine biosynthesis, catabolism and transport [19].
- Glycine riboswitches bind glycine to regulate glycine metabolism genes, including the use of glycine as an energy source. As of 2008, this riboswitch is the only known natural RNA that exhibits cooperative binding [R3].
- cyclic di-GMP riboswitches bind the signaling molecule cyclic di-GMP in order to regulate a variety of genes controlled by this second messenger [11].
- GlcN6P riboswitch is a ribozyme that cleaves itself when there is a sufficient concentration of glucosamine-6-phosphate [20].
A small RNA regulator typically can be turned on by transcriptional controls or turned off by an enzyme that degrades the RNA regulator product. RNAs are usually thought to be unable to have allosteric properties, and thus not expected to respond to small molecules by changing its ability to recognize the target. Riboswitches appear to be the exception. Specific, high affinity binding of the ligand stabilizes one particular three-dimensional conformation of the riboswitch, leading to the formation of a stem-loop structure. If no ligand is bound, a different three-dimensional conformation of the riboswitch becomes energetically more favorable and is adopted. Where this stem-loop forms, the characteristics of the region involved determine the overall effect of the binding, of which three main mechanisms exist:
- formation of stem-loops that lead to transcription attenuation (occlusion of RNA polymerase)
- inhibition of translation initiation (exclusion of ribosome at Shine-Dalgarno site, or RBS)
- cleavage of mRNA
Different mechanisms in the functioning of the same riboswitch have been observed. For example, in B. subtilis, the RFN element, binding flavin mononucleotide (FMN) acts on the rib operon mRNA via transcription termination but acts on the ypaA transcript via sequestration of ribosome binding sequences.
An example mechanism of action for a self-cleaving riboswitch is shown for the regulation of the system that produces the metabolite GlcN6P [8, 20]. In this case, GlcN6P binds to the catalytic site on the riboswitch and activates its ability to cleave the RNA in an intramolecular reaction [R4, R5].

Figure Credit: Roxana Ordoñez, Adapted from [8]
Of the riboswitch structures that have been solved, two types of structures have emerged. Type I riboswitches are comprised of a preorganized tertiary complex with a single catalytic pocket. Purine riboswitches, the glmS riboswitch, and the SAM-II riboswitch are of this type. Type II riboswitches are made up of a two party catalytic pocket whose tertiary structure is induced by ligand binding. This type includes the TPP riboswitch, the SAM-I riboswitch, and the M-box magnesium riboswitch [R3]. All known riboswitches fold into compact RNA secondary structures with a base stem, a central multi-loop and several branching hairpins.
Riboswitches are sequences of RNA located in the 5’ untranslated region of mRNAs that can fold complex structure and serve as receptors of specific metabolites, thus enabling the regulation of that gene’s expression levels. Acting as an aptamer, they preferentially bind to their specific metabolite, undergoing conformational changes allosterically and regulating the process of transcription and/or translation [9]. Thus, the two key characteristics of riboswitches that distinguish them from other ribozymes or RNAs are 1) their ability to directly bind a metabolite to RNA in the absence of proteins, and 2) gene regulation is dependent on the binding of said metabolite. The figure below shows a cartoon of a riboswitch bound to its target molecule and a potential expression platform.

Figure Credit: Roxana Ordoñez
- Coppins et al. The intricate world of riboswitches. Curr Opin Microbiol (2007) vol. 10 (2) pp. 176-81
- Henkin. Riboswitch RNAs: using RNA to sense cellular metabolism. Genes Dev (2008) vol. 22 (24) pp. 3383-90
- Montange and Batey. Riboswitches: emerging themes in RNA structure and function. Annual review of biophysics (2008) vol. 37 pp. 117-33
- Serganov and Patel. Ribozymes, riboswitches and beyond: regulation of gene expression without proteins. Nat Rev Genet (2007) vol. 8 (10) pp. 776-90
- Serganov. The long and the short of riboswitches. Curr Opin Struct Biol (2009) vol. 19 pp. 1-9
- Barrick and Breaker. The power of riboswitches. Sci Am (2007) vol. 296 (1) pp. 50-7
- Bauer and Suess. Engineered riboswitches as novel tools in molecular biology. J Biotechnol (2006) vol. 124 (1) pp. 4-11
- Bengert and Dandekar. Riboswitch finder--a tool for identification of riboswitch RNAs. Nucleic Acids Res (2004) vol. 32 (Web Server issue) pp. W154-9
- Blount et al. Antibacterial lysine analogs that target lysine riboswitches. Nat Chem Biol (2007) vol. 3 (1) pp. 44-9
- Gesteland et al. The RNA World. (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, ed. 3, 2006), pp. 89–107
- Grundy and Henkin. The S box regulon: a new global transcription termination control system for methionine and cysteine biosynthesis genes in gram-positive bacteria. Mol Microbiol (1998) vol. 30 (4) pp. 737-49
- Isaacs et al. Engineered riboregulators enable post-transcriptional control of gene expression. Nat Biotechnol (2004) vol. 22 (7) pp. 841-7
- Lewin. Genes IX. (Jones and Bartlett Publishers, Inc. 2008), pp. 260-261
- Mandal and Breaker. Gene regulation by riboswitches. Nat Rev Mol Cell Bio (2004) vol. 5 (6) pp. 451-63
- Sudarsan et al. Metabolite-binding RNA domains are present in the genes of eukaryotes. RNA (2003) vol. 9 (6) pp. 644-7
- Sudarsan et al. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science (2008) vol. 321 (5887) pp. 411-3
- Vitreschak et al. Riboswitches: the oldest mechanism for the regulation of gene expression?. Trends Genet (2004) vol. 20 (1) pp. 44-50
- Winkler et al. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature (2002) vol. 419 (6910) pp. 952-6
- Winkler et al. An mRNA structure that controls gene expression by binding FMN. Proc Natl Acad Sci USA (2002) vol. 99 (25) pp. 15908-13
- Winkler and Breaker. Regulation of bacterial gene expression by riboswitches. Annu Rev Microbiol (2005) vol. 59 pp. 487-517
- Nahvi et al. Coenzyme B12 riboswitches are widespread genetic control elements in prokaryotes. Nucleic Acids Res (2004) vol. 32 (1) pp. 143-50
- Roth et al. A riboswitch selective for the queuosine precursor preQ1 contains an unusually small aptamer domain. Nat Struct Mol Biol (2007) vol. 14 (4) pp. 308-17
- Mandal et al. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell (2003) vol. 113 (5) pp. 577-86
- Sudarsan et al. An mRNA structure in bacteria that controls gene expression by binding lysine. Genes Dev (2003) vol. 17 (21) pp. 2688-97
- Winkler et al. Control of gene expression by a natural metabolite-responsive ribozyme. Nature (2004) vol. 428 (6980) pp. 281-6
- Win and Smolke. A modular and extensible RNA-based gene-regulatory platform for engineering cellular function. Proc Natl Acad Sci USA (2007) vol. 104 (36) pp. 14283-8
Since riboswitches are found in mRNAs, their biogenesis would follow accordingly. In short, mRNAs are produced by transcription, where a single-stranded RNA identical in sequence with one of the strands of the duplex DNA is generated. Transcription occurs in three stages: initiation, elongation, and termination. RNA polymerase initiates transcription after binding to a promoter site on DNA. During elongation, the newly synthesized RNA chain is extended as the polymerase moves along the DNA. Termination occurs when the polymerase and RNA are released and the DNA duplex reforms [8]. Since riboswitches have been observed primarily in prokaryotes, riboswitches can be assumed to generally be transcribed without the more elaborate post-transcriptional processing seen in eukaryotes. However, riboswitches have been found in simpler eukaryotes, indicating their potential for splicing controls [10]. For more information, please see the wiki page on mRNA.
|
Group |
Members | Natural Ligand | Size (nucleotides) | Distribution |
| Coenzymes | TPP (also THI-box) | TPP, thiamine pyrophosphate | 100 | Bacteria, archaea, eukaryotes (fungi, plants) |
| FMN (also RFN-element) | FMN, flavin mononucleotide | 120 | Bacteria | |
| AdoCbl (also B12-element) | AdoCbl, adenosylcobalamin | 200 | Bacteria | |
| SAM-I (also called S-box) | SAM, S-adenosylmethionine | 105 | Mostly Gram+ bacteria | |
| SAM-II | SAM, S-adenosylmethionine | 60 | α- and β-proteobacteria | |
| SAM-III (SMK) | SAM, S-adenosylmethionine | 80 | Gram– bacteria | |
| Amino Acids | Lysine (also L-box) | Lysine | 175 | γ-proteobacteria, Thermotogales, Firmicutes |
| Glycine (I+II) | Glycine | 110 | Bacteria | |
| Nucleobases | Guanine (also G-box) | Guanine, hypoxanthine | 70 | Gram+ bacteria |
| Adenine | Adenine | 70 | Bacteria | |
| preQ1 | preQ1, pre-quenosine-1 | 35 | Bacteria | |
| Self-cleaving mRNA | glmS | GlcN6P, glucosamine-6-phosphate | 170 | Gram+ bacteria |
Adapted from [R4].
Some mRNA regulatory elements, such as thermosensor RNA and T-box RNA, share features of riboswitches. Whether they belong to the riboswitch class or not is still unclear, as is the distinction between riboswitches and other gene control systems. Although the number of riboswitch classes that exist varies, there are more than 200 unique riboswitches that have been categorized into about 9-12 classes [R4].
Some examples of secondary structure for a few riboswitches:
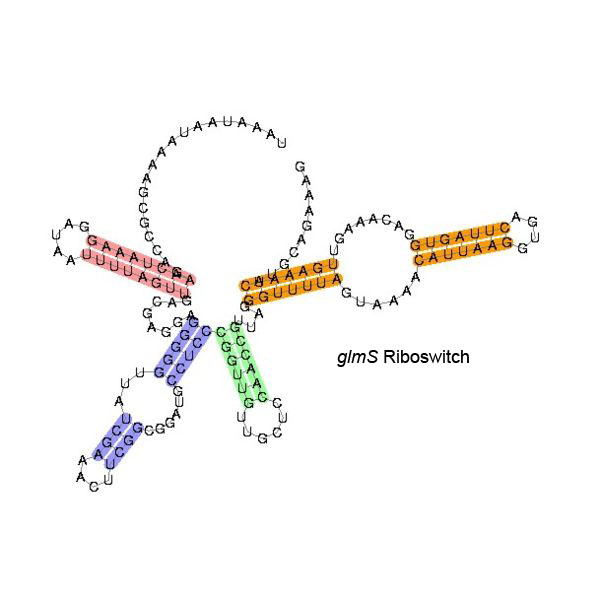

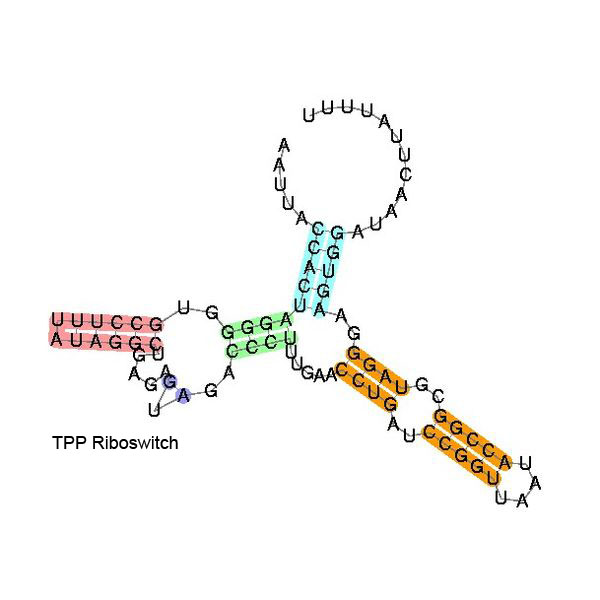
The secondary structure images are taken from the Rfam database, which is completely in the public domain.The right to use this work is granted to anyone for any purpose, without any conditions, unless such conditions are required by law.
In addition to their potential as anti-bacterial targets, riboswitches have recently become popular in the field of synthetic biology. Designer riboswitches could be customized to bind nearly any molecule of choice to affect a gene’s expression level [1, 2, 7]. In 2007, Christina Smolke designed and developed an entirely modular RNA-based gene-regulatory device [21]. In this modular and extensible system, a library of aptamers could be inserted developed around the hammerhead ribozyme. Whereas naturally occurring riboswitches are highly selective and specifc, this modular platform provides a universal system in which the ligand, aptamer, and expression platform are customizable.
Prior to the naming of the riboswitch, Tina Henkin and coworkers may have been the first to observe this special gene control system. They found that there was a conserved sequence in the 5’ UTR of the S-box gene family involving the biosynthesis of methionine and cysteine [6]. Computer predictions showed that this conserved sequence formed secondary structure or possibly tertiary structure, and combined with experimental results, Henkin showed that this structure would change when the leader region was bound to a negative regulatory factor.
In 2002, Breaker first demonstrated that mRNAs can bind metabolites directly in the absence of proteins. Further, his group also developed a useful method to detect the conformational change of mRNA. He found that the conformational change of the riboswitch in the btuB gene, whose product is a membrane protein for the transportation of coenzyme B12, is regulated by coenzyme B12 (AdoCbl) with high specificity [13, 14].
Since Breaker’s discovery, researchers have revisited experimental data now known to correspond to riboswitch function dating back at least 30 years. Several previously labeled gene regulation 'mysteries' can now be explained by the presence of riboswitches [9]. Computational predictions have allowed for uncovering of numerous novel RNA motifs and small non-coding RNAs expressed by bacteria, suggesting that more riboswitches will be identified. Further, these findings indicate a significant role for gene regulation by riboswitches in bacteria.
Currently, the majority of riboswitches studied have been in bacteria, however recent studies have found functional riboswitches in some eukaryotes (i.e. plants and certain fungi) [10, 11].
While there is no known role for riboswitches in human disease, Breaker and coworkers have demonstrated that lysine analogs that bind to lysine riboswitches have antibacterial activity. These findings hold promise for powerful approaches to bacterial drug resistance [4]. Further, engineered control of gene expression could be applied in the context of gene therapy, where the riboswitch could function as a highly specific sensor for a benign drug-like molecule [1].

