Guide RNA directed insertion/deletion RNA editing is phenomena unique to the mitochondria of kinetoplastid trypanosomes. The current hypothesis is that this highly specialized molecular mechanism evolved in the early ancestral bodonid lineage after spitting off from the Euglenoid lineage. (reviewed in 15)
Genetic variation among Tryoanosomatids is encoded not within the genomic sequences of protein encoding genes found in the highly conserved maxicircles but within the heterogeneity of an individual specie’s gRNA, which direct the editing of these mRNA sequences (9).
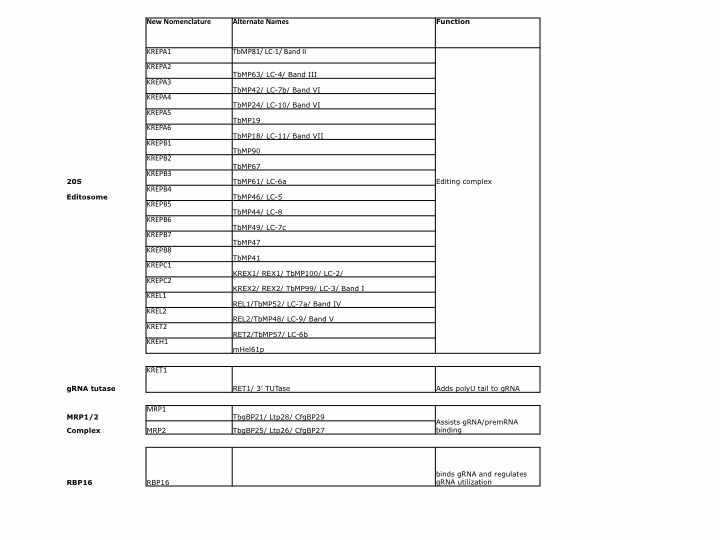
gRNA interact directly with components of the insertion/deletion 20S editosome as well as several RNA binding accessory factors as discussed above. The nomenclature for the various protein components of the Trypanosome editing machinery has become very complicated due to the fact that several labs have described what turned out to be the same factors in the same organism or analogous factors in different Trypanosome species. Below is a summary table of these factors with the new nomenclature introduced by Stuart et. al, 2005 (9) and Lukes et al. 2005 (16).
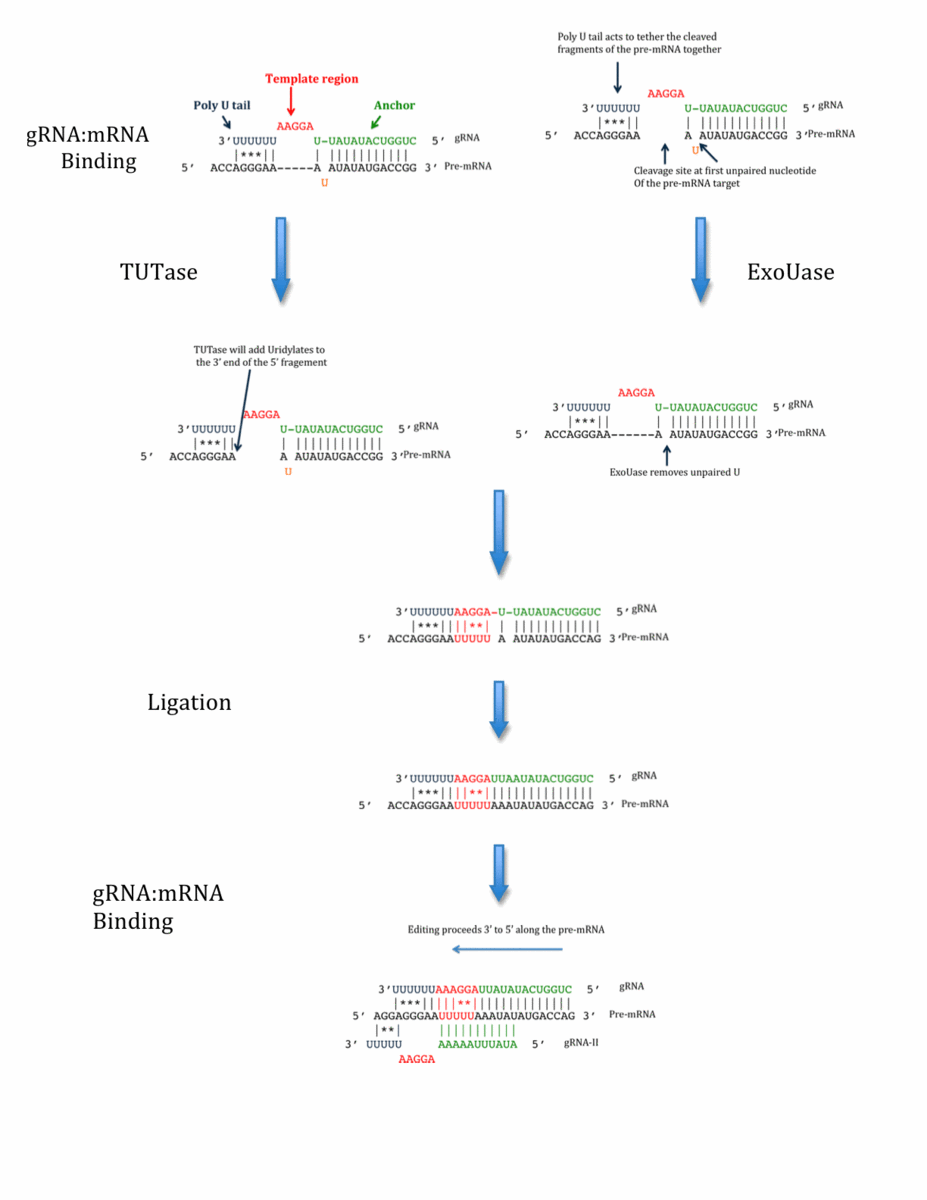
The currently accepted mechanism of gRNA directed RNA editing is the enzymatic cascade model originally proposed by Blum et. al. in 1990. Editing occurs in a 3’ to 5’ direction and is thought to occur in 4 stages: gRNA/mRNA anchor duplex formation, cleavage of the mRNA target, the addition or deletion of U residues in the mRNA and finally the re-ligation of the two fragments of the newly edited mRNA.
gRNA/mRNA anchor duplex formation
The 5’ ends of gRNAs contain 10-15 nucleotides called the anchor region and it is this region that is directly complementary to a target unedited mRNA or pre-mRNA. The double stranded region formed by the binding of the gRNA anchor with its target pre-mRNA is called the anchor duplex and it forms directly 3’ to the region of the pre-mRNA to be edited. Anchor duplex formation brings the 3’ poly U tail of the gRNA into close proximity with purine rich regions of the pre-mRNA. This interaction may serve to stabilize the gRNA/pre-mRNA duplex during the subsequent editing steps and results in a bulging mismatch region of the gRNA that serves as an information template for U addition/deletion. The pairing of a gRNA with its pre-mRNA target is directed by the MRP1/2 protein complex (12).
Cleavage
The endonucleolytic cleavage of the pre-mRNA occurs at the first unpaired nucleotide directly 3’ (with respect to the gRNA) of the gRNA/mRNA anchor duplex. Interactions between the poly U tail of the gRNA and purine rich regions of the pre-mRNA are thought to tether together the 5’ and 3’ fragments of the cleaved pre-mRNA. The 20S editosome, a protein complex containing all of the enzymatic machinery necessary to catalyze editing, contains seven proteins with predicted endonucleases motifs, however, the specific role each plays in the pre-mRNA cleavage is not yet understood. In addition it has been suggested that distinct endoribonucleases may catalyze cleavage depending upon whether insertion or deletion editing is occurring. (13)
Addition/deletion of Uridylates
Depending on the information encoded in the template region of the gRNA Uridylates may either be added to the pre-mRNA by a Terminal Uridyl transferase (TUTase) alternately Uridylates may be deleted by an Uridyl exonuclease (exoUase).
Cleavage of the pre-mRNA results in the production of a hydroxyl group on the 3’ end of the 5’ fragment. In the process of U addition, free UTP moieties are added to this hydroxyl group by the editosome TUTases KRET2. In deletion editing one or both of the exoUases KREX1 and KREX2 cleave unpaired U in the pre-mRNA in a 3’ to 5’ direction.
Re-ligation
Following U insertion or deletion the 5’ and 3’ fragments of the pre-mRNA are ligated through the action of the editosome RNA ligases KREL1 and KREL2 (14). Although these two ligases are very similar in sequence they may play slightly different roles. KREL1 appears to be necessary for both insertion and deletion editing where KREL2 appears to be necessary only for insertion editing.
In kinetoplastid Trypanosomes guide RNAs (gRNAs) direct the insertion and or deletion of uridylates in mitochondrial mRNA. gRNA directed mRNA editing is necessary for the maturation and proper function of 12 out of 18 mitochondrial protein encoding mRNAs (9). Not surprisingly, proper gRNA function is essential for full completion of the Trypanosome life cycle, as organisms that have impaired or absent gRNA function do not survive past the procyclic or insect stage of development (3).
Guide RNA directed editing normally is thought to correct frameshift errors present in the gene sequence present in the mitochondrial genome as originally described by Benne. Editing of the cytocrome b gene, however, results not in a frameshift but the generation of a start codon (10) demonstrating that gRNA directed editing can correct multiple types of genomic sequence errors.
Additionally, several recent studies have demonstrated that guide RNA directed editing might be a mechanism by which Trypanosomes may regulate the functional diversity of their protein encoding genes. Recently it was shown that alternate editing of the coxIII gene leads to a novel function of this gene (11). This report has lead to the speculation that guide RNA directed editing in trypanosomes like alternate splicing in eukaryotes may be a mechanism to encode for protein diversity and evolutionary adaptation.
Lukes J, Hashimi H, Zíková A. Unexplained complexity of the mitochondrial
genome and transcriptome in kinetoplastid flagellates. Curr Genet. 2005
Nov;48(5):277-99
Simpson L, Thiemann OH, Savill NJ, Alfonzo JD, Maslov DA. Evolution of RNA
editing in trypanosome mitochondria. Proc Natl Acad Sci U S A. 2000 Jun
20;97(13):6986-93
Stuart KD, Schnaufer A, Ernst NL, Panigrahi AK. Complex management: RNA
editing in trypanosomes. Trends Biochem Sci. 2005 Feb;30(2):97-105
1. Benne R, Van den Burg J, Brakenhoff JP, Sloof P, Van Boom JH, Tromp MC. Major
transcript of the frameshifted coxII gene from trypanosome mitochondria contains
four nucleotides that are not encoded in the DNA. Cell. 1986 Sep 12;46(6):819-26
2. Blum B, Bakalara N, Simpson L. A model for RNA editing in kinetoplastid
mitochondria: "guide" RNA molecules transcribed from maxicircle DNA provide the
edited information. Cell. 1990 Jan 26;60(2):189-98
3. Hashimi H, Cicová Z, Novotná L, Wen YZ, Lukes J. Kinetoplastid guide RNA
biogenesis is dependent on subunits of the mitochondrial RNA binding complex 1
and mitochondrial RNA polymerase. RNA. 2009 Apr;15(4):588-99
4. Pollard VW, Rohrer SP, Michelotti EF, Hancock K, Hajduk SL. Organization of
minicircle genes for guide RNAs in Trypanosoma brucei. Cell. 1990 Nov
16;63(4):783-90
5. Grams J, McManus MT, Hajduk SL. Processing of polycistronic guide RNAs is
associated with RNA editing complexes in Trypanosoma brucei. EMBO J. 2000 Oct
16;19(20):5525-32
6. Aphasizhev R, Aphasizheva I, Simpson L. A tale of two TUTases. Proc Natl Acad
Sci U S A. 2003 Sep 16;100(19):10617-22
7. McManus MT, Adler BK, Pollard VW, Hajduk SL. Trypanosoma brucei guide RNA
poly(U) tail formation is stabilized by cognate mRNA. Mol Cell Biol. 2000
Feb;20(3):883-91
8. Ochsenreiter T, Cipriano M, Hajduk SL. KISS: the kinetoplastid RNA editing
sequence search tool. RNA. 2007 Jan;13(1):1-4
*9. Stuart KD, Schnaufer A, Ernst NL, Panigrahi AK. Complex management: RNA
editing in trypanosomes. Trends Biochem Sci. 2005 Feb;30(2):97-105
10. Feagin JE, Shaw JM, Simpson L, Stuart K. Creation of AUG initiation codons by
addition of uridines within cytochrome b transcripts of kinetoplastids. Proc Natl
Acad Sci U S A. 1988 Jan;85(2):539-43. Erratum in: Proc Natl Acad Sci U S A 1988
May;85(10):3540
11. Ochsenreiter T, Cipriano M, Hajduk SL. Alternative mRNA editing in
trypanosomes is extensive and may contribute to mitochondrial protein diversity.
PLoS ONE. 2008 Feb 13;3(2):e1566
12. Müller UF, Lambert L, Göringer HU. Annealing of RNA editing substrates
facilitated by guide RNA-binding protein gBP21. EMBO J. 2001 Mar
15;20(6):1394-404
13. Cruz-Reyes J, Rusché LN, Sollner-Webb B. Trypanosoma brucei U insertion and U
deletion activities co-purify with an enzymatic editing complex but are
differentially optimized. Nucleic Acids Res. 1998 Aug 15;26(16):3634-9
14. McManus MT, Shimamura M, Grams J, Hajduk SL. Identification of candidate
mitochondrial RNA editing ligases from Trypanosoma brucei. RNA. 2001
Feb;7(2):167-75
*15. Simpson L, Thiemann OH, Savill NJ, Alfonzo JD, Maslov DA. Evolution of RNA
editing in trypanosome mitochondria. Proc Natl Acad Sci U S A. 2000 Jun
20;97(13):6986-93
*16. Lukes J, Hashimi H, Zíková A. Unexplained complexity of the mitochondrial
genome and transcriptome in kinetoplastid flagellates. Curr Genet. 2005
Nov;48(5):277-99
gRNAs are 45-70nt RNAs transcribed from primarily from the minicircles of the mitochondrial genome of Kinetoplastid Trypanosomes. Minicircles contain on average between 1-4 gRNAs and the transcription of gRNA occurs most likely through the action of the mitochondrial polymerase (3)which also transcribes the protein encoding mRNAs of the maxicircles. It is believed that gRNA genes contain their own transcriptional start sites as gRNA transcription usually begins at a conserved 5’-RYAYA-3’ sequence and gRNAs contain a 5’ triphosphate (4). Interestingly each minicircle seems to have only a single transcriptional termination sequence and as a result gRNA’s are often transcribed as polycistrons from which only the 5’ most gRNA is properly processed (potentially by the editosome) while the downstream transcripts are degraded (5).
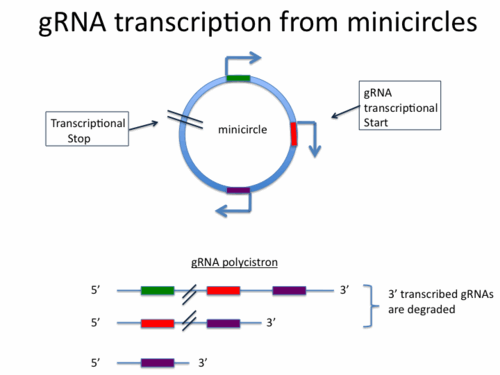
gRNA’s themselves are post-transcriptionally modified by the addition of a poly U tail of variable lengths. It has been proposed (6 ) that the TUT II complex containing the KRET1 TUTase is responsible for this process. Additionally it has been proposed that poly U tail formation may be stabilized through binding of purine rich regions in the target RNA. (7)
Because the genomes of some Kinetoplastids contain upwards of 10,000 gRNAs, many of which are species specific, the use of computerized databases is a useful tool for those studying gRNAs. One such tool is the KISS or the Kinetoplastid Insertion and deletion sequence search tool ( http://rna.bmb.uga.edu/kiss/ ) (8) . KISS is a web-based database of known and predicted gRNAs for T. Brucei which also contains known and predicted mRNA targets for different gRNAs. Another useful resource is the U-insertion/deletion Edited Sequence Database ( http://dna.kdna.ucla.edu/trypanosome/database.html ) which has sequence information of both gRNA’s and their mRNA targets from several Trypanosome species.
Due to the fact that guide RNAs are unique to Trypanosomes their use as a biological tool may be restricted to researchers who study these organisms. Perturbing gRNA function could result in altered or absent mRNA function although such an approach would likely be less effective than RNAi based strategies, which have been shown to be effective in several Trypanosome species.
Guide RNA also know as gRNA are responsible for directing U insertion/deletion editing in Kinetoplastid Trypanomes.
The concept of RNA editing proposed by Benne et al in 1986 (1) challenged the central dogma of molecular biology by demonstrating the genetic information encoded in mRNA can vary from that encoded in the genome. These authors showed that the not only does the mature mitochondrial mRNA sequence encoding the critcical coxII gene in kinetoplastid trypanosomes contain 4 Uridylate bases not encoded in the genome but that the addition of these U residues is necessary to correct a frameshift in the genomic sequence. Subsequent work demonstrated that a majority of the Kinetoplastid genome requires insertion/deletion editing to produce functional transcripts. Four years after Benne’s initial description, in 1990 Blum et. al. (2) identified the RNA species that direct or guide the posttranscriptional modification of mRNA sequences and coined the term guide RNA or gRNA .
Trypanosmes are exclusively parasitic protozoa and are the causative agent in several human diseases including African sleeping sickness, Chagas Disease and the Leishmaniases. Although gRNA itself is not pathogenic they are essential to kinetoplatid survival and thus may represent a potential therapeutic target.

