RNase P catalyzes the endonucleolytic 5' cleavage of tRNA precursors in all of the kingdoms of life (Archaea, Bacteria and Eukarya) and in mitochondria and chloroplasts. While the RNA subunit has been conserved among the kingdoms, Bacteria have recruited different protein from Archaea and Eukarya.
RNA component:
The RNA subunit of RNase P of Bacteria, members of Archaea and Eukarya shares some structural homology. Bacterial RNA subunits are 350-450 nucleotides in length and are made up of two types of RNA that are classified based on their secondary structure: A and B type. The B type evolved from the A type and differs in the number of structural elements attached to the homologous structures that are conserved across species. A similar sized subunit is found in Archaea and Eukarya. Within this region there are five critical regions (CRI-V) which are the homologous conserved regions in bacterial, archaeal and eukaryotic nucelar RNase P RNAs (6).
Protein components:
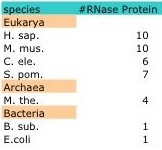
Despite the structural homology evident among members of the three kingdoms, the protein compositions varies greatly. Bacterial RNase P have only one protein moiety that is not required for catalytic function of the RNA subunit in vitro (5,15). However, members of the Archaea and Eukarya contain multiple protein subunits which are indispensible for in vitro function of the catalytic RNA subunit. For example, Methanothermobacter thermoautotrophicus contain four protein subunits while S. cerevisiae have nine and Homo sapiens ten. While overlapping proteins can be found within and among the Archaea and Eukarya, no evolutionary relationship has been found between the bacterial proteins and any of the archaeal and eukaryotic RNase P proteins (15).
In bacteria, RNase P is small ribonucleoprotein complex composed of a catalytic RNA and a protein cofactor. In E.coli, the protein subunit (C5) is not essential for the catalytic function of the RNA subunit (M1 RNA) in vitro (3). In vivo, C5 helps modulate sustrate affinity and specificity to the C domain of M1 RNA; however, bacterial RNase P is a ribozyme with M1 RNA carrying out the catalytic activity of cleaving pre-tRNA. Archaeal and Eukaryal RNase P are larger ribonucleoprotein complexes with as many as ten proteins associated with the RNA subunit (Table). Archaeal and Eukaryal RNase P are composed of proteins that share homology and themselves function as enzymes to help catalyze the cleavage of the pre-tRNA (9).
Table 2: Number of RNase P protein in the three kingdoms.
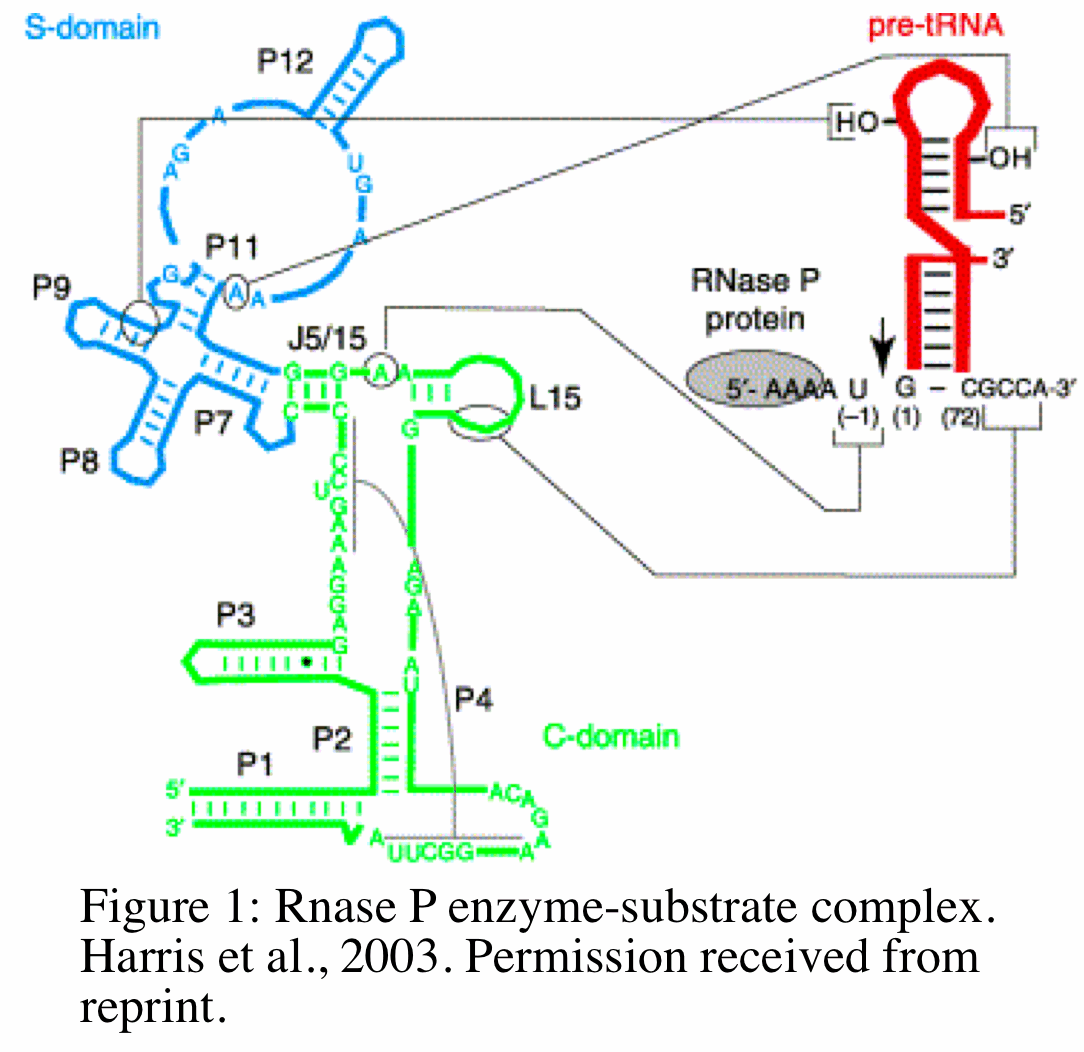 RNase P functions as a endoribonuclease that removes extraneous 5' sequences from tRNA precursor by catalyzing the hydrolysis of a specific phosphodiester bond. The mechanism by which this occurs in bacteria has been studied extensively and reveals unique and equally important functions for the RNA and protein subunits. The RNA subunit (M1 RNA) of bacterial RNase P binds to pre-tRNA at two of its domains: the specificity domain (S domain) and catalytic domain (C domain) (Figure 1,18). The C domain includes the ribozyme active site that interacts with the 3' sequence of pre-tRNA. The protein subunit (C5) of bacterial RNase P binds (at a specific cleft of the protein) to pre-tRNA at the 5' leader sequence (grey circle in Figure). The position and orientation of the the protein- pre-tRNA binding places the protein and the 5' leader sequence of pre-tRNA in close proximity to the ribozyme active site within the C-domain (18). The M1 RNA and C5 protein work together to catalyze the cleavage of pre-tRNA at the C domain and help guide specific substrates to the active site of the C domain, respectively. In addition to modulating substrate specificity, recently Kim et al. 2009 showed in E.coli that the protein subunit is required for the metabolic stability of M1 RNA in vivo. Without the protein subunit, M1 RNA is metabolically unstable and will degrade with a half life of 5 minutes in vivo (8).
RNase P functions as a endoribonuclease that removes extraneous 5' sequences from tRNA precursor by catalyzing the hydrolysis of a specific phosphodiester bond. The mechanism by which this occurs in bacteria has been studied extensively and reveals unique and equally important functions for the RNA and protein subunits. The RNA subunit (M1 RNA) of bacterial RNase P binds to pre-tRNA at two of its domains: the specificity domain (S domain) and catalytic domain (C domain) (Figure 1,18). The C domain includes the ribozyme active site that interacts with the 3' sequence of pre-tRNA. The protein subunit (C5) of bacterial RNase P binds (at a specific cleft of the protein) to pre-tRNA at the 5' leader sequence (grey circle in Figure). The position and orientation of the the protein- pre-tRNA binding places the protein and the 5' leader sequence of pre-tRNA in close proximity to the ribozyme active site within the C-domain (18). The M1 RNA and C5 protein work together to catalyze the cleavage of pre-tRNA at the C domain and help guide specific substrates to the active site of the C domain, respectively. In addition to modulating substrate specificity, recently Kim et al. 2009 showed in E.coli that the protein subunit is required for the metabolic stability of M1 RNA in vivo. Without the protein subunit, M1 RNA is metabolically unstable and will degrade with a half life of 5 minutes in vivo (8).
In addition to its endoribonuclease function, human RNase P has also been found to act as a transcription factor also plays a role in transcription through association with chromatin of tRNA genes. Seven of the ten proteins tested so far have coimmunoprecipitated with the chromatin of tRNA genes. The protein subunits associate with tRNA genes in dividing HeLa cells and dissociate from the chromatin in cells that cease to divide. It is possible that the presence of RNase P during transcription of tRNA is an example of coordination of transcription and regulation; however, coimmunoprecipitation experiments revealed that active RNase P protein can be pulled down with Pol III from genes that are not known to be substrates of RNase P (5S rRNA, 7SL RNA and U6 snRNA genes) suggesting that RNase P is acting as a transcription factor (7).
RNase P functions as an endoribonuclease that removes extraneous 5' sequences from precursor tRNAs to generate mature tRNA (2, 17). In 2007, 36 years after the discovery of RNase P and its catalytic function, Jarrous et al. found a novel function for human nuclear RNase P. By chromatin immunoprecipitation, the authors found that human RNase P is necessary for transcription of small noncoding RNA genes (tRNA, rRNA) by Pol III in whole HeLa cells and cell extracts (7).
15. Hartmann E, Hatmann R. (2003) The enigma of ribonuclease P evolution. TRENDa in Genetics. 19: 561-569. PMID: 14550630
16. Altman S. (2007) A view of RNase P. Mol. BioSyst. 3:604-607. PMID: 17700860
17. Altman S. (1975) Biosynthesis of transfer RNA in Escherichia coli. Cell. 4:21–29. PMID: 1090377
18. Harris M, Christian E. (2003) Recent insights into the structure and function of the ribonucucleoprotein enzyme ribonuclease P. Curr Opin Struct Biol. 13:325-333. PMID: 12831883 Permission recieved for reprint of figure.
19. Kim K, Liu F. (2007) Inhibition of gene expression in human cells using RNase P-derived ribozymes and external guide sequences. Biochim Biophys Acta. 1769;603-12. PMID: 17976837
1. Altman S. (1971) Isolation of tyrosine tRNA precursor molecules. Nat. New Biol. 229:19-21. PMID: 5277330
2. Altman S, Smith JD. (1971) Tyrosine tRNA precursor molecule polynucleotide sequence. Nat. New Biol. 233:35-39. PMID: 4938965
3. Robertson HD, Altman S, Smith JD. (1972) Purification and properties of a specific Escherichia coli ribonuclease which cleaves a tyrosine transfer ribonucleic acid presursor. J Biol Chem. 247:5243–5251. PMID: 4560501
4. Stark B, Kole R, Bowman E, Altman S. (1978) Ribonuclease P: An enzyme with an essential RNA component. Proc. Natl. Acam. Sci. 75: 3717-3721. PMID: 358197
5. Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. (1983) The RNA moiety of Ribonuclease P is the catalytic subunit of the enzyme. Cell. 35:849-857. PMID: 6197186
6. Chen JL, Pace NR. (1997) Identification of the universally conserved core of ribonuclease P RNA. RNA 3: 557–560. PMID: 9174091
7. Reiner R, Ben-asouli Y, Krilovetzky I, Jarrous N. (2006) A role for the catalytic ribonucleoprotein RNase P in RNA polymerase III transcription. Genes Dev. 20:1621–1635. PMID: 16778078
8. Kim Y, Lee Y. (2009) Novel funtion of C5 protein as a metabolic stabilizer of M1 RNA. FEBS Lett. 538: 419-424. PMID: 19114042
9. Hall TA, Brown JW. (2002) Archaeal RNase P has multiple protein subunits homologous to eukaryotic RNase P proteins. RNA. 8:296-306. PMID: 12003490
10. Liu F, Altman S. (1995). Inhibition of viral gene expression by the catalytic RNA subunit of RNase P from Escherichia coli. Genes Dev. 9:471-480. PMID: 7533740
11. Cobaleda C, Sanchez-Garcia I. (2000) In vivo inhibition by a site-specific RNA catalytic RNA subint of RNase P designed aagainst the BCR-ABL oncogenic products: a novel approach for cancer treatment. Blood. 95: 731-737. Blood Journal
12. Guerrier-Takada C, Li Y, Altman S. (1995) Artificial regulation of gene expression in Escherichia coli by RNase P. Proc Natl Acad Sci U S A. 92: 11115–11119. PMID: 7479948
13. Kraus G, Geffin R, Spruill G, Young A, Seivright R, Cardona D, Burzawa J, Hnatyszyn HJ. (2002) Cross-clade inhibition of HIV-1 replication and cytopathology by using RNase P-associated external guide sequences.Proc Natl Acad Sci U S A. 99: 3406–3411. PMID: 11904403
14. Haifeng Zhang H, Altman S. (2004) Inhibition of the Expression of the Human RNase P Protein Subunits Rpp21, Rpp25, Rpp29 by External Guide Sequences (EGSs) and siRNA. 342:1077-1083. PMID: 15351636
The Evolution of RNA and Protein Subunit(s): RNase P is found in living organisms across all three kingdoms. The homologues of the RNA subunits across the phylogenetic domains suggests that they descended from a common progenitor. However, the number and structure of protein subunit in Bacteria compared to Archaea and Eukarya differ. This evolutionary enigma that continues to puzzle scientists. One hypothesis proposes that RNA evolved first and when Bacteria and Archaea diverged, they evolved different methods for recruiting protein subunit(s) (15).
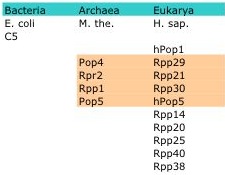
RNase P is a member of a class of nucleases called ribonucleases that function to catalyze the cleavage of RNA. However, RNase P is unique from its family members; it is a ribozyme made of one (Bacteria) or more (Archaea, Eukarya ) protein moieties and a catalytic RNA moiety whose enzymatic activity is thought to be inversely proportional to the number of protein subunits (5, 16). While the RNA subunit is conserved, the number of protein subunits and their structure varies across species with some conservation seen among species within the Archaea and Eukarya. For example, Escherichia coli contain only one protein subunit, Methanothermobacter thermoautotrophicus four and Homo Sapiens ten with homology seen in four of the proteins between M. the. and H. sap (Table1).
Table 1: Example members of the RNase P protein subunit. Four highlighted protein in M. the. are orthologs of the four highlighted proteins in H.sap. (15)
Gene Therapy: Even without its protein subunit, bacterial M1 RNA is highly efficient at cleaving pre-tRNA in vitro. This ability to cleave pre-tRNAs makes Bacterial RNase P a useful gene therapy tool for the destruction of bacterial and viral mRNAs (10, 19) and oncogenes (11,19). Guide sequences (GS) are designed as complementary sequences to the target mRNA and contain an unpaired 3' CCA sequence. The complementarity of the GS allows for hybridization to the target mRNA and the 3' unpaired sequences allows for recognition and cleavage by M1 RNA. The GS hybridized to its target mimics an M1 RNA natural substrate that is recognized by M1 RNA and subject to destruction. The GS can be external (EGS) and independent of the M1 RNA or internal (IGS) and covalently linked to the 3' end of the M1 RNA (refered to as M1GS RNA) (10, 19). Guerrier-Takada et al. 1995 showed specific targeting of EGS to B-galactosidase and alkaline phosphatase encoding genes whose expressions were decreased by 50-60% in E.coli suggesting that the carefully designed EGS was successful at recruiting endogenous RNase P (12). This study revealed the antibacterial application of EGS technology to specifically inhibit expression of bacterial genes. Around the same time, another group showed successful use of IGS for blocking viral replication. Liu et al. showed that an IGS fused to M1 RNA can specifically bind mRNA encoding thymidine kinase (TK) of herpes simplex virus 1 (HSV-1). Mouse cells in culture expressing the M1GS RNA construct and infected with HSV-1 showed an ~80 percent reduction in TK mRNA and protein levels (10). In addition to HSV-1, RNase P and M1 RNA have been used as an antiviral agent by many laboratories to block infection of human immunodeficiency virus, human influenza virus, human cytomegalovirus and Kaposi's sarcoma-associated herpesvirus (19). Studies have also used EGS as a antiviral agent for the successful recruitment of endogenous RNase P in targeted cleavage and degradation of viral mRNA. Kraus et al. used an EGS against the U5 region of HIV-1 mRNA to recruit endogenous RNase P for the destruction of HIV-1 mRNA. The authors were able to show that T cells expressing the EGS targeting HIV-1 mRNA and challenged with HIV did not produce significant levels of HIV throughout the 30 day infection period (13). This result provided the first evidence that RNase P-associated EGSs may serve as potential anti-HIV agents.
In addition to the anti-bacterial and anti-viral potential of RNase P, studies also show that RNase P can be used as a technology to target cancer. Cobaleda et al. used chronic myelogenous and acute lymphoblastic leukemia models where abberant translocation results in BCR-ABL oncogenes. Using M1GS RNA that targeted the oncogenic mRNA at the fusion site, the authors were able to successful block the effect of BCR-ABL function in cultured mammalian cells (11).
Basic Science Research: M1GS and EGS have possible use in basic science research as a tool for understanding cellular function by suppressing gene expression. However, in recent years, the use of RNA interference to degrade mRNA has been the focus for gene interference studies because of its powerful effect with small concentration and ability to use cellular machinery to knockdown mRNA. In an attempt to compare the activity and effectiveness of EGS vs. siRNA technology, Zhang et al. showed that the inhibition of RNase P protein subunits by siRNA was comparable to EGS. The time it took to see the inhibition took longer with siRNA technology (48-96 hours post transfection) as compared to EGS technology (24 hours) (14). Additionally, since EGS technology depends on the catalytic actions of RNase P, an abundant, stable and efficient enzyme, cleavage of target mRNA is highly efficient and irreversible. Scientists have also found little cytotoxicity in cells expressing these molecules (19).
RNase P is the short name for ribonucleus P. RNase P is found in species across all kingdoms and is made up of one RNA subunit and one (Bacteria) or many (Archaea and Eukarya) proteins. The bacterial RNase P RNA has two classes based on secondary structure including A- type (ancestral-type) and B-type (Bacillus-type). RNase P of Escherichia coli is composed of an A-type RNA referred to as M1 RNA and one protein subunit referred to as C5. Bacillus subtilis contains RNase P RNAs that are B-type. Human RNA is referred to as H1 RNA and the ten protein subunits include hPop1, Rpp29, Rpp21, Rpp30, hPop5, Rpp14, Rpp20, Rpp25, Rpp40, Rpp36 (Table1). In addition to these eleven protein subunits, there are four others (Pop6, Pop3, Pop8, Pop7) that are found in yeast (15).
Discovery and Purification of RNase P:
While studying mutants of tyrosine tRNA in E.coli, Sidney Altman observed that his mutants were not only longer than the wildtype but they reverted to the wildtype at a high rate. In 1971, Altman isolated the transcript of the gene for tyrosine tRNA and found that there were extra nucleotides at the 5' and 3' ends of what would become recognized as the precursor tRNA molecule (1). Isolation of this precursor revealed a 129 nucleotide long tRNA species that contains 44 extra nucleotide sequences in addition to the 85 already present in mature tRNA. Of the 44, 41, including a 5' terminal nucleoside triphosphate, are found on the 5' end of the mature tRNA. Altman and Smith showed that mixing the precursor tRNA of 129 nucleotides long with an E.coli extract resulted in the cleavage of 5' and 3' ends of the precursor (2, 17). This discovery lead them to the hypothesis that specific ribonucleases must be processing precursor molecules giving rise to functional tRNA. And indeed in 1972, Robertson, Altman and Smith purified a specific ribonuclease from E.coli extracts leading to the discovery of RNase P. They found that RNase P cleaves a single phosphodiester bond of the 129 nucleotide tyrosine tRNA precursor which removes extra nucleotides present at the 5' terminus of the precursor. However, they did not find an effect upon the extra nucleotides at the 3' end of the tRNA precursor hypothesizing that another ribonuclease must be present for cleaving the 3' end (3).
RNase P: A protein-RNA mix
In 1978, under the mentorship of Sidney Altman, Benjamin Stark discovered that purified RNase P from E.coli is made up of one RNA species and one discrete protein species. Stark analyzed purified RNase P on a polyacrylamide and showed one prominent band staining with Coomassie brilliant blue (protein stain) and one band staining with methylene blue (nucleic acid stain). The bouyant density of RNase P in CsCl further revealed the presence of RNA and predicted a 1:4 ratio of protein to RNA (4).
RNase P: a ribonuclear ribozyme
In 1983, Cecilia Guerrier-Takada working in Sidney Altman's laboratory found that the RNA subunit of RNase P alone is sufficient to carry out catalytic cleavage of tRNA precursor in high concentrations of magnesium ions. The enzymatic property of the RNA subunit independent of the protein distinguishes RNase P from other ribonucleases as it is one of two ribozymes in nature (5).

