All life forms, with the exception of prions, requires mRNA synthesis to direct new protein production. Viruses must have their genetic information, whether it is stored as DNA or RNA, converted to an mRNA-like entity capable of being read by the host ribosomes. Individual mRNA transcripts vary in proportion to the genetic diversity of organisms and their propensity for alternative splicing. Messenger RNA corresponding to highly conserved genes will be similarly conserved between species. Human β-actin cDNA, made by reverse transcribing mRNA, was sequenced and found to maintain 79% sequence identity with the rat cDNA in coding regions (Ponte et al 1984). All changes were silent, such that the amino acid sequences of the transcribed mRNAs were identical. Less, but significant, conservation was observed in the untranslated regions (UTR). Also, conservation between chicken and human 3’ UTR was observed. These data suggest that coding sequences are generally more highly conserved among species than the untranslated regions.
https://www.ncbi.nlm.nih.gov/sites/entrez (pre-mRNA and processed mRNA transcripts of desired genes can be obtained)
Messenger RNA pairs with many proteins and other RNAs during its existence. At first, pre-mRNA synthesis is catalyzed by RNA polymerase. It then interacts 7-methyl guanylyl transferase and other capping enzymes, poly(A) polymerase, and with the splicing machinery of the cell, which is made up of both proteins and small nuclear RNA. Interactions with the nuclear porin complex mediates transfer of the mRNA from the nucleus to the cytoplasm. In the cytoplasm, the mRNA binds to the ribosomal subunits, which include rRNA, and also to translation initiation factors and tRNA. Translation may be attenuated by the binding of micro RNAs to the mRNA.
The poly(A) tail can act independently of the cap to recruit ribosomes for translation of the mRNA into a protein. Preiss et al found that the extent of this recruitment correlated with the length of the tail and suggested that poly(A) tail length might be a mechanism for controlling the extent of translational for an individual mRNA (1998). This may be mediated by poly(A) binding protein (Pab1p), which could also associate with the cap binding protein eIF4G. eIF4G might then recruit other ribosomal machinery.
Once recruited and assembled, the ribosome begins to translate the mRNA to amino acids. The three base codon is the operational unit of RNA and each codon guides the incorporation of one of the twenty amino acids to the growing C-terminal peptide. Recognition of the codon is directly mediated by a complementary anti-codon on an amino acid – conjugated transfer RNA (tRNA). The genetic code has redundancy, as 64 possible codons code for only 20 amino acids (and start and stop). Evolution appears to have selected for similarities between codons encoding the same amino acids such that a group of codons differing only at the third base will encode the same amino acid (Novozhilov et al 2007). This provides robustness to the translation process; 69% of mutations at the third residue of a codon are synonymous, and mispairings at that codon between the mRNA and tRNA have reduced odds of leading to a mutated protein. Furthermore, amino acid residues with similar molecular characteristics also bear some similarities with respect to their codons.
The Genetic Code:
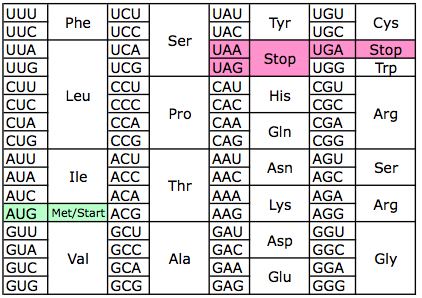
Three letter codes in caps represent the three nucleotides, from 5’ to 3’, that make up each particular codon. These are followed by the encoded amino acid. Codons specifying the same amino acid are contained in the same box.
In the cytoplasm of eukaryotes, mRNA serves as a template for ribosomal synthesis of new proteins. Messenger mRNA is read from 5’ to 3’. At either end of coding mRNA lies untranslated regions which contain sequences regulating the efficiency of location of translation. mRNA stability and synthesis are important areas for regulation of gene expression levels.
Douglas L. Black. Mechanisms of Alternative Pre-Messenger RNA Splicing. Annu. Rev. Biochem. 2003. 72: 291–336 PMID: 12626338
Hahn S. Structure and mechanism of the RNA polymerase II transcription machinery. 2004. Nature Structure and Molecular Biology 11: 394-403. PMID: 15114340
Kapp LD, Lorsch JR. The Molecular Mechanisms of Eukaryotic Translation. 2004. Annual Review of Biochemistry 73: 657-704. PMID: 15189156
Brenner S, Jacob F, Meselson M. An unstable intermediate carrying information from genes to ribosomes for protein synthesis. 1961. Nature 190: 576-81.
Darnell JE, Philipson L, Wall R, Adesnik M. Polyadenylic Acid Sequences: Role in Conversion of Nuclear RNA into Messenger RNA. 1971. Science 174: 507-10. PMID: 5110429
Dintzis HM. Assembly of the Peptide Chains of Hemoglobin. 1961. PNAS 47: 247-262. PMID: 13723017
Goldstein L, Plaut W. Direct Evidence for Nuclear Synthesis of Cytoplasmic Ribose Nucleic Acid. 1955. PNAS 41: 874-880. PMID: 16589764
Hess PR et al. Vaccination with mRNAs encoding tumor-associated antigens and granulocyte-macrophage colony-stimulating factor efficiently primes CTL responses, but is insufficient to overcome tolerance to a model tumor/self antigen. 2006. Cancer Immunol Immunother 55: 672-83. PMID: 16133108
Jensen KB, Dredge BK, Stefani G, Zhong R, Buckanovich RJ, Okano HJ, Yang YY, Darnell RB. Nova-1 Regulates Neuron-Specific Alternative Splicing and Is Essential for Neuronal Viability. 2000. Neuron 25: 359-71. PMID: 10719891
McCracken S et al. 5’ Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. 1997. Genes and Development 11:3306-3318. PMID: 9407024
Miller OL, Hamkalo BA, Thomas CA. Visualization of Bacterial Genes in Action. 1970. Science 169: 392-5. PMID: 4915822
Novozhilov AS, Wolf YI, and Koonin EV. Evolution of the genetic code: partial optimization of a random code for robustness to translation error in a rugged fitness landscape. 2007. Biology Direct 2: 1-24. PMID: 17956616
Phillips AV, Cooper TA. RNA Processing and Human Disease. 1999. CMLS 57: 235-49. PMID: 10766020
Ponte P et al. Evolutionary conservation in the untranslated regions of actin mRNAs: DNA sequence of a human beta-actin cDNA. 1984. Nucleic Acids Res 12: 1687-96. PMID: 6322116
Preiss T, Muckenthaler M, and Hentze MW. Poly(A)-tail-promoted translation in yeast: implications for translational control. 1998. RNA 4: 1321-31. PMID: 9814754
Sammeth M, Foissac S, Guigo R. A General Definition and Nomenclature for Alternative Splicing Events. 2008. PLOS Comput Biol 4: 1-14. PMID: 18688268
Volkin E, Astrachan L. Phosphorus Incorporation in Escherichia coli Ribonucleic Acid after Infection with Bacteriophage T2. 1956. Virology 2: 149-161.
Ule J, Stefani G, Mele A, Ruggiu M, Wang X, Taneri B, Gaasterland T, Blencowe BJ, Darnell RB. An RNA map predicting Nova-dependent splicing regulation. 2006. Nature 444: 580-6. PMID: 17065982
West S, Proudfoot NJ, Dye MJ. Molecular Dissection of Mammalian RNA Polymerase II Transcriptional Termination. 2008. Molecular Cell. 29: 600-10. PMID: 18342606
Transcription
An RNA polymerase synthesizes mRNA from DNA in both prokaryotes in eukaryotes. Prokaryotes have only one RNA polymerase, while eukaryotes have three. Prokaryotic transcription appears to be guided to promoter primarily by the sigma subunit of the complex. In eukaryotes, RNA polymerase II (pol II) binds to promoters of transcribed genes with the aid of several general transcription factors and is responsible for synthesizing mRNA (Hahn 2004). In both prokaryotes and eukaryotes, binding of the basal transcription machinery is affected by gene specific factors that may act both by inducing chromatin changes in the DNA and by directly recruiting of the polymerase complex. The pre-initiation complex has formed when all required components are assembled. This is followed by a shift to the open complex in which about 15 bp of DNA is unwound. Transcription initiation is followed by elongation. In eukaryotes, pol II has a C-terminal domain with repeated YSPTSPS motifs. The serines in these motifs are sites of regulatory phophorylations, and can serve as binding sites for various factors mediating transcription processes. Elongation is then followed by termination, a multi-step process that involves release of the transcript from the template DNA followed by processing of the poly(A)-tail (West et al 2008). Transcriptional termination occurs at specific sequences; both the poly(A) signal and downstream termination signals contribute.
Processing
Maturing mRNAs are modified by the addition of a 5’ cap and a poly(A) 3’ tail. The poly(A) tail addition occurs independent of mRNA synthesis, and is added in the nucleus prior to export to the cytoplasm (Darnell et al 1971). Inhibition of poly(A) tail synthesis reduces mRNA presence in the cytoplasm, suggesting that the poly(A) tail mediates stability of the pre-mRNA/hnRNA. 5’ capping enzymes responsible for adding a 7-methyl guanosine to the end of the pre-mRNA are recruited by the C-terminal domain of RNA pol II (McCracken et al 1997). The 5’ cap then facilitates splicing, export, and translation of the mRNA.
Splicing
Exons of about 50-300 nucleotides encode sequence information that eventually determine the primary amino acid sequence of newly synthesized proteins (Douglas 2003). These are separated by longer introns (insert more info on introns). Non-coding introns in pre-mRNA are excised by a complex of protein and RNA called the spliceosome. Exonic splicing enhancers and silencers encoded in the pre-mRNA affect the sites of splicing. Enhancer sites are recognized by Serine/Arginine rich (SR) proteins (Douglas 2003), which then mediates recruitment of spliceosome components. Conversely, hnRNPs associate at silencer sites and inhibit splicing. Binding of hnRNPs to silencer sites leads to an excision of the silenced exon when the preceding and following exons are spliced together. In addition to these exonic enhancers and silencers, regulatory elements capable of directing the spliceosome machinery are also present in introns.
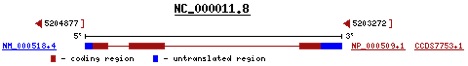
Figure 1. A diagram of the human hemoglobin beta gene and the corresponding mRNA produced. Transcription of the entire gene is followed by splicing out of the thinner red lines, which represent introns. The bolder red regions represent exons that are spliced together as pre-mRNA is processed to mature mRNA. Blue regions are also part of the final transcript and control translation and mRNA stability.
Alternative Splicing
The splicing of the same pre-mRNA into different transcripts is one means by which higher organisms can increase the diversity of their proteome while maintaining fairly consistent numbers of genes. Different protein isoforms can arise from the same gene by differential inclusion of introns. Up to 60% of human genes could be alternatively spliced (Douglas 2003). Most exons are constitutive and will always be included in the final mRNA. Cassette exons are either entirely present or absent in the final mRNA, while a third subset of exons may have some of their sequence excluded through alternative splicing.
The protein Nova controls tissue-specific splicing events in the mammalian brain and illustrates how alternative splicing can generate increased diversity of the proteome. Nova-1 binds to conserved sites in both introns and exons, and the presence of these sites has been used to map the probable splicing events affected by Nova (Ule et al 2006). The phenotype of the Nova deficient mouse highlights the important of these splicing events in vivo; the mouse dies shortly after birth from motor defects (Jensen et al 2000).
Messenger mRNA exists for any transcribed gene, of which there are over 20,000 in the human genome. Futhermore, alternative splicing creates multiple mRNA transcripts from these genes. The β-globin mRNA has been well studied (West et al 2008).
Injection of messenger RNA encoding tumor antigens to mice as a means of immunization against tumor growth has been investigated, and investigators have also used cDNA injections as a vaccination tool. Injection of dendritic cells transfected with antigen-coding mRNAs appears to have some efficacy in vivo, but direct injection of mRNA appears to be less effective at breaking tolerance to tumors (Hess et al 2006).
Messenger RNA is usually abbreviated as mRNA. In the nucleus, pre-mRNA is transcribed, and this precursor RNA makes up the majority of heterogenous nuclear RNA (hnRNA). After splicing and nuclear export to the cytoplasm it is called mRNA, and is then considered to be mature. Particular mRNAs can be referred to by the protein that they encode. Alternatively splice mRNAs can be referred to by the protein’s particular isoform, and newer classification systems based on the splicing events conducted have also been devised (Sammeth et al 2008).
The first description of mRNA was made by Elliot Volkin and Lazarus Astrachan in 1956, although its role was not appreciated (Volkin et al 1956). These investigators found that infection of E. coli with bacteriophage led to the synthesis of a new RNA species distinct in its base composition. It was later shown that the base composition of this new minor RNA species mirrored that of the bacteriophage. In 1961, it was appreciated that ribosomes were the site of protein synthesis and that DNA stored genetic material. The method of protein synthesis was unclear, as was the mediator that carried the information from DNA to the ribosomes. However, Lester Goldstein and Walter Plaut had radiolabeled RNA and observed its synthesis in the nucleus prior to movement to the cytoplasm in 1955 (Goldstein et al 1955). Sydney Brenner, Francois Jacob, and Matthew Meselson conducted experiments to show that phage infection was followed by synthesis of new RNA but not ribosomes, thus discounting the hypothesis that individual ribosomes might be made specifically for each protein (Brenner et al 1961). In 1961, it was also discovered that protein synthesis occurred in a linear manner from the N to C terminus of the protein. Reticulocytes, isolated for their almost exclusive synthesis of hemoglobin, were shown to incorporate new radiolabeled nucleotides only at the C-terminal end of newly formed hemoglobin (Dintzis et al 1961). Finally, the function of mRNA was more firmly establish in 1970 when mRNA coming off of extruded nuclear DNA was shown to be bound directly by multiple ribosomes (Miller et al 1970).
General defects in RNA processing pathways are not observed, because these mutations are embryonic lethal. However, a few mutations affecting splicing only a subset of genes have been described (Phillips et al 1999). Also, individual genes may be inappropriately spliced when mutated, and the microtubule-binding protein tau is one such example. Changes in splicing of CD44 is associated with the acquisition of metastatic potential by human tumors.

