The two snoRNA guide families (box C/D and box H/ACA) have been identified in multiple eukaryotic species including metazoans as well as yeast, plants, and kinetoplastid protozoans. In bacteria rRNA nucleotide modifications are present (four 2'-O-methylations and ten pseudouridines), however these were found to be catalyzed by site specific protein enzymes without any additional RNA cofactor. Interestingly, snoRNA guides to direct nucleotide modifications have also been found in archaea. Not only do these similar mechanisms exist in archaea and eukaryotes, but archaeal homologs of both box C/D and H/ACA snoRNAs have been identified suggesting a these RNA modifications found in eukaryotes may have its ancestral origins in archaea rather than bacteria (Bachellerie et al. 2002).
It has been proposed that the H/ACA snoRNP classes originally evolved from a gene duplication of a tRNA specific pseudouridine synthase, that later acquired the capacity to recognize an ancestral H/ACA snoRNA as well as the pre-rRNA rather than a tRNA. One potential candidate for the ancestral snoRNA is snR30, which is involved in pre-rRNA cleavage, rather than modification. This snoRNA over time through duplications and random mutations would have given rise to snoRNAs with novel complementarity to rRNA sequences to direct modification of new sites (Lafontaine et al. 1998). A similar evolutionary progression has been proposed for the box C/D snoRNAs that direct 2'-O-methylation of rRNA nucleotides. The primordial box C/D snoRNA is thought to be U3, which like snR30, is not involved in nucleotide modification, but rather is required for cleavage and processing of rRNA (Lafontaine et al 1998).
Human snoRNA Database
Yeast snoRNA Database
The plant snoRNA database
Rfam database of RNA families
snoRNA Wikipedia Page
In vertebrates the C/D box snoRNAs associate with four other protein partners to generate the functional snoRNPs. These include fibrillarin, NOP56, NOP65/NOP58, and 15.5kDa. Fibrillarin is the methyltransferase enzyme that is directly responsible for the additional of a methyl group to the 2'-O of the target nucleotide and the 15.5 kDa protein component has been implicated in binding to the C/D motif. Together this complex guides the methylation of rRNA nucleotides (Filipowicz et al. 2002).
The H/ACA box snoRNAs in vertebrates also interact with a core set of four proteins. These include dyskerin, GAR1, NHP2, and NOP10. Dyskerin is the pseudouridine synthase which catalyzes the conversion of uridine to psuedouridine and GAR1 and NHP2 have been implicated in RNA binding. Homologous protein complexes have also been identified in yeast and other eukaryotes.
Additional protein components have been shown to interact with the snoRNP complexes to assist with RNP biogeneiss. p55 and p50 proteins have been suggested to have RNA helicase activity while others like SMN and Nopp140 have suggested roles in SNP assembly and SNP trafficking, respectively (Filipowicz et al. 2002).
2'-O-Methylation
Box C/D snoRNAs, that direct the 2'-O-methylation of rRNA nucleotides contain are defined by two short sequence motifs after which they are named. Box C (5'RUGAUGA3') is found a few nucleotides away from the 5' end of the snoRNA while box D (5'CUGA3') is only a few nucleotides away from the opposite 3' end. The 4-5 nucleotides at both termini of the snoRNA come together to form a terminal stem-box structure which has been shown to be required for snoRNA biogenesis as well as nucleolar localization (Bachellerie et al. 1998). Another less conserved C box, termed C', is found in the central portion of the snoRNA, while another D box, termed D', is located in the 5' region. It is in these regions that RNA-protein interactions occur to direct the proper assembly of the functional ribonucleoprotein complexes (Kiss 2001).
Upstream of the box D or D' elements there are either one or two antisense elements. These are sequences of 10-21 nucleotides that are complementary to a specific site of rRNA that will allow for proper alignment and methylation of the appropriate nucleotide. These complementary stretches form double helical structures with the corresponding rRNA sequences. The location of the D or D' box predicts the exact location of the nucleotide to be modified, as it is always the fifth position upstream from this box element onto which the methyl group is added. While the snoRNA component directs the snoRNP complex to the appropriate rRNA location, it is a protein enzyme that actually catalyzes the methylation reaction. In the case of the box C/D snoRNPs, it is the associated protein fibrillarin that is the methyltransferase (Bachellerie et al. 2002).
Pseudouridylation
Box H/ACA snoRNAs form a secondary structure made up to two large hairpin domains connected by a hinge region, followed by a short tail. The conserved box motifs include the H (5'ANANNA3' where N is any nucleotide) and the ACA trinucleotide always found three nucleotides away from the 3' end of the snoRNA. The H box is found in the hinge region, while the ACA motif is in the tail region. Guide sequences that direct the snoRNA to the appropriate rRNA sequence are found in one or both of the hairpin loop domains. After targeted the snoRNP complex to the appropriate site, the substrate rRNA uridine is located at the base of the upper stem where it closes the recognition loop of the snoRNA (Ganot et al 1997). As with the box C/D snoRNAs, the location of the H or ACA box of the snoRNA in relation to the uridine to be modified is important in determining the correct pseudouridylation site. Generally there is 14-16 nucleotide distance between the box motifs and the site to be modified by the pseudouridine synthase, which in humans is called dyskerin (Kiss 2001).
To summarize, for both of the classes of snoRNAs, it is the snoRNA itself that targets the snoRNP complex to the appropriate site in ribosomal RNA, where a protein emzyme present in the complex catalyzes the nucleotide modification.
RNA Modification
SnoRNAs are involved in guiding the modification of rRNA nucleotides. During ribosome biosynthesis, the pre-rRNAs must undergo several modifications mainly 2'-O-ribose methylation and pseudouridylation (See below). More specifically, the human 18S, 5.8S, and 28S rRNAs cumulatively contain approximately 110 2'-O-methyl groups and almost 100 pseudouridines (Maden et al, 1990).
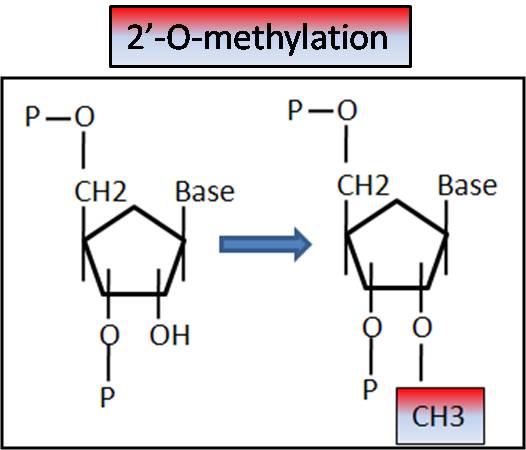

In order to properly modify rRNA nucleotides, snoRNAs are assembled into small nucleolar ribonuloprotein particles which include the nucleotide modifying enzymes (either a methyltransferase, or a pseudouridine synthase). Based on sequence complementarity, the snoRNA components of these snoRNPs direct the nucleotide modifying enzymes to the appropriate rRNA sequence (Kiss, 2001). While nucleotide changes directed by snoRNAs are generally seen to be dispensable for cell growth and viability, they are likely involved in refining RNA-RNA as well as RNA-protein interactions. The 2'-O methylation of nucleotides may protect the RNA from hydrolytic degradation, enhance hydrophibic surfaces for interaction, or stabilize helical stems. Pseudouridines have increased flexibility in their C-C glycosyl bonds and therefore may allow for increased capacity for hydrogen bond formation, contributing to RNA tertiary structure (Bachellerie et al. 2002).
While snoRNAs were originally described as rRNA modifying enzymes, their cellular functions continue to expand. It has now been demonstrated that snoRNAs also are able to modify small nuclear or snRNAs that mediate mRNA splicing. There are five major spliceosomal RNAs found in mammals: U1, U2, U4, U5, and U6. Together these five snRNAs have 30 2'-O-methylations and 24 pseudouridines (Reddy et al. 1998). SnoRNA guides complementary to these snRNA sites have been identified and in some cases been shown to be required for the nucleotide modifications, though in some cases it appears that of the pseudouridines are generated by protein enzymes (Huttenhoger et al 2001). One unique snoRNA, U85, is a hybrid box C/D-H/ACA, and has been shown to guide the 2'-O-methylation of C45 and the pseudouridylation of U46, two neighboring nucleotides, in the U5 snRNA (Jady et al 2001).
SnoRNAs may also play a role in modifying mRNAs. One example was found in a brain specific box C/D snoRNA that contains an 18 nucleotide conserved target recognition element that is 100 percent complimentary tot he serotonin receptor 5-HT(2C) mRNA. Interestingly, the putative methylation site that would be modified by this snoRNA is known to undergo adenosine to inosine editing. This may raise the possibility that snoRNA guided 2'-O-methylation may also contribute to control of expression of this brain specific protein (Cavaille et al. 2000). There are also an increasing number of so called "orphan" snoRNA guides identified using bioinformatic approaches to which no complementary sequence can be found (Kawaji et al. 2008). With new discoveries for cellular RNA functions increasing, it is possible that novel roles for snoRNA mediated nucleotide modifications have yet to be discovered.
RNA Processing
Some snoRNAs are involved in the processing of pre-rRNAs rather than nucleotide modification. These include C/D RNAs U3, U8, U14, and U22, as well as H/ACA RNAs snR10, snR30, E2 and E3. These snoRNAs direct processing machinery to specific cleavage sites on pre-rRNAs (Venema et al. 1999; Kressler et al. 1999).
(1) Lafontaine, DLJ. Tollervey, D. 1998 "Birth of the snoRNPs: the evolution of the modification-guide snoRNAs." Trends Guide to Bioinformatics p. 383-388 PMID: 9810226
(2) Olson, Mark OJ. 2004. The Nucleolus. Published by Springer 2004.
(3) Maxwell, ES. Fournier, MJ. 1995. "The Small Nucleolar RNAs." Annual Reviews in Biochemistry. 35:897-943. PMID: 7574504
(4) Kawaji, H. Hayashikazi, Y. 2008. "Exploration of Small RNAs." PLoS Genetics. Vol. 4, Issue 1. p3-8. PMID: 18225959
(5) Chanfreau G, Rotondo G, Legrain P, Jacquier A. 1998 "Processing of a dicistronic small nucleolar RNA precursor by the RNA endonuclease Rnt1." EMBO J, 17:3726-3737. PMID: 9649442
(6) Allmang C, Kufel J, Chanfreau G, Mitchell P, Petfalski E, Tollervey D. 1999. "Function of the exosome in rRNA, snoRNA and snRNA synthesis." EMBO J 1999, 18:5399-5410. PMID: 10508172
(7) van Hoof A, Lennertz P, Parker R. 2000. "Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs." Mol Cell Biol 2000, 20:441-452. PMID: 10611222
(8) Terns MP, Terns RM. 2002. "Small nucleolar RNAs: versatile trans-acting molecules of ancient evolutionary origin." Gene Express 10:17-39. PMID: 11868985
(9) Weinstein LB, Steitz JA. 1999. "Guided tours: from precursor snoRNA to functional snoRNP." Curr Opin Cell Biol 1999, 11:378-384. PMID: 10395551
(10) Maden, BEH et al. 1990. "The numerous modified nucleotides in eukaryotic ribosomal RNA." Prog. Nucleic Acid Res. Mol. Biol. 39:241-303. PMID: 2247610
(11) Reddy, R. Busch, H. 1988. Small nuclear RNAs: RNA sequences, structure and modifications. In Birnstiel, M.L. "Structure and Function of Major and Minor Small Nuclear Ribonucleoprotein Particles." Springer-Verlag, Berlin, Germany. p1-37.
(12) Huttenhofer, A et al. 2001. "RNomics: an experimental approach that identifies 201 candidates for novel, small non-messenger RNAs in mouse." EMBO J. 20:2943-2953. PMID: 11387227
(13) Jady, BE. Kiss, T. 2001. "A small nucleolar guide RNA functions both in 2'-O-methylation and pseudouridylation of the U5 splieosomal RNA." EMBO J. 20:541-551. PMID: 11157760
(14) Cavaille, J. et al. 2000. "Identification of brain specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization." Proc. Natl. Acad. Sci. USA, 97:14311-14316. PMID: 11106375
(15) Venema J, Tollervey D. 1999. "Ribosome synthesis in Saccharomyces cerevisiae." Annu Rev Genet 33:261-311. PMID: 10690410
(16) Kressler D, Linder P, de la Cruz J. 1999 "Protein trans-acting factors involved in ribosome biogenesis in Saccharomyces cerevisiae." Mol Cell Biol, 19:7897-7912. PMID: 10567516
(17) Bachellerie, JP. Cavaille, J. 1998. "Small nucleolar RNAs guide the ribose methylation of eukaryotic rRNAs." In Modification and Editing of RNA: The Alteration of RNA Structure and Function. ASM Press, Washington, DC. p255-272.
(18) Ganot, P et al. 1997. "Site specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs." Cell. 89:799-809. PMID: 9182768
(19) Samarsky, DA et al. 1999. A small nucleolar RNA: ribozyme hybrid cleaves a nucleolar RNA target in vivo with near-perfect efficiency,
Proc. Natl. Acad. Sci. USA 96:6609–6614. PMID: 10359759
(20) Nicholls, RD. Knepper, JL. 2001. "Genome organization, function, and imprinting in Prader-Willi and Angelman syndromes." Annu. Rev. Human Genet. 2:153–175 PMID: 11701647
Gene Structure and Transcription
 Eukaryotic cells exhibit differences in both the genetic organization and mechanisms of biosynthesis of snoRNAs. While only a few vertebrate snoRNAs involved in rRNA processing (not modification), such as U3 or U8 are trancsribed as independent genes with their own promoter nearly all snoRNAs identified in yeast are generated in this manner. On the other hand, plants often generate polycistronic transcripts from which multiple snoRNAs are processed. In vertebrates, all of the snoRNAs that are known to guide nucleotide modifications are located within introns of genes transcribed via RNA polyermase II (See Figure). Many snoRNAs are embedded within introns of genes that themselves are involved in ribosome biogenesis (Terms et al. 2002, Steitz et al 1999).
Eukaryotic cells exhibit differences in both the genetic organization and mechanisms of biosynthesis of snoRNAs. While only a few vertebrate snoRNAs involved in rRNA processing (not modification), such as U3 or U8 are trancsribed as independent genes with their own promoter nearly all snoRNAs identified in yeast are generated in this manner. On the other hand, plants often generate polycistronic transcripts from which multiple snoRNAs are processed. In vertebrates, all of the snoRNAs that are known to guide nucleotide modifications are located within introns of genes transcribed via RNA polyermase II (See Figure). Many snoRNAs are embedded within introns of genes that themselves are involved in ribosome biogenesis (Terms et al. 2002, Steitz et al 1999).
snoRNA Processing
Once snoRNAs have been transcribed if they are in polycistronic units or introns, they need to be processed to produce the mature RNA sequence that can then assemble properly into the the small nucleolar ribonucleoprotein particles and guide the modification or cleavage of rRNA nucleotides. In yeast, pre-snoRNAs are removed from polycistronic transcripts through the action of the endoribonuclease Rnt1p, which cleaves RNA within specific hairpin structures (Rotondo et al. 1998). Once each snoRNA has been removed, the newly formed ends are furthered processed by 5' and 3' specific exonucleases to remove any remaining nucleotides not part of the mature snoRNA (Allmang et al, 1999; van Hoof et al. 2000). Intronic snoRNAs in vertebrates are generally liberated from introns through canonical splicing, however there have been reports suggesting some are removed by specific endonucleases as is seen in the yeast polycistronic trancsripts. After liberation from introns, these snoRNAs are processed to remove excess nucleotides from either end via exonuclease activity as seen in yeast (See below). Signal sequences within the snoRNAs centered at boxes C and D or H and ACA directs binding of protein interacting partners that represent the functional snoRNP complex. Interaction and assemble of these protein-RNA complexes are required for the proper processing and localization of snoRNAs (Terns et al 2002; Weinstein et al 1999).

There are currently greater than 200 snoRNAs identified in mammals, the majority of which direct the methylation (box C/D) or pseudouridylation (box H/ACA) of rRNA nucleotides. One chimerican example, U85, has been identified that contains features of both of the snoRNA classes and can alone mediate the addition of both of these modifications.
There are a few snoRNAs however, that do not function to guide nucleotide modifications. These snoRNAs are required for directed cleavage of pre-rRNAs during processing. Some notable examples are listed below.
Box C/D members: (human) U3, U8, U14, U22
Box H/ACA members: (human) U17, E2, E3 (yeast) snR10, snR30
SnoRNAs have been exploited as an experimental tool in two different ways. First, snoRNA localization elements have been utilized to direct new RNA sequences to the nucleolus. For example, the C/D box structural motif was added to a chimeric hammerhead ribozyme, which was then successfully targeted to the nucleolus was it was shown to efficiently cleave nucleolar RNA selectively (Samarsky et al. 1999).
Second, snoRNAs have been used to direct nucleotide modifications to new sites in rRNA via the expression of snoRNAs containing novel guide sequences. Engineered antisense elements in snoRNAs have successfully generated novel methylation and pseudouridylation sites on target rRNAs. Additionally, this same strategy has proven effective at mediating site-directed modification of other RNA polymerase II and III transcripts (Bachellerie et al 2002). This provides a tool for researchers to explore the role of RNA modifications in various cellular processes.
Small nucleolar RNAs, abbreviated snoRNAs, are divided into two groups. Based on sequence motifs and secondary structures they are classified as either Box C/D or Box H/ACA snoRNAs, which are involved in the methylation and pseudouridylation of rRNA nucleotides, respecitvely (Lafontaine et al. 1998). Many of the vertebrate snoRNAs are named beginning with a U designation. This designation emerged after the initial purification of snoRNA species, in which they were shown to possess a high uridine content, leading to the naming convention beginning with U with a number following, though this is not uniform across all of the identified vertebrate snoRNAs (Olson 2004). In yeast, snoRNAs are named systematically starting with lower case 'sn' followed by capital 'R' and then an identifying number.
The designation snoRNA was coined in 1981, but the initial identification of the first small nucleolar RNA, U3, occurred over ten years earlier. Upon fractionation of RNA from rat cell nuclei via gel electrophoresis, additional RNA bands were observed tha represented the snRNA and snoRNA species. As these were found to be rich in uridine, the U designation used today was created (Olson, 2004). The history of snoRNA research can be divided into two general time periods. This initial period of discovery in the late 1960s and early 1970s encompassed not only the discovery of snoRNAs but also demonstrated their association with ribosomal RNAs leading to their predicted function in ribosome synthesis, a hypothesis that was strengthened after demonstrating chemical crosslinking of U3 snoRNA to rRNA. Research in this field then turned to the identification of other snoRNAs, or U3 variants, as well as characterizing U3 snoRNA gene structure, expression, and structural analysis of the U3 RNA and snoRNP complexes (Maxwell et al. 1995). Upon sequencing several U3 snoRNAs, conserved box sequences (C/D) and related secondary structure were found. Box C/D were later identified in other snoRNAs, as well as another distinct box H/ACA element on several other species, leading to the current two class designation of snoRNAs (Olson 2004). These efforts fell under the broader theme of exploring the pathway of rRNA processing.
In the late 1980s, a new phase of snoRNA research began. This new interest in ribosome synthesis, snoRNAs and novel RNA functions was aided by simultaneous advances in new strategies and technologies for analyzing these processes. Antibodies specific for trimethylguanosine (the product of snoRNA modification of rRNA) as well as the nucleolar protein fibrillarin (a component of a snoRNP complex) aided in snoRNA detection and allowed for separation from other nucleoplasmic RNAs. Since then, the understaning of snoRNA structure and function has greatly expanded.
Currently, new snoRNAs are generally identified via bioinformatic approaches looking for sequence complimentarity between snoRNA and rRNA or other RNA sequences. These new broader screening techniques have not only led to the identification of a huge number of snoRNAs is various organisms, but also found a number of "orphan" snoRNA guides in which no known RNA targets can be identified. These snoRNAs but ultimately lead to the discovery of novel cellular functions for these RNAs (Kawaji et al. 2008).
One orphan snoRNA, HBII-52 is located in an imprinted locus and expressed in the brain, has been shown to mediate alternative splicing of the serotonin receptor 2C by utilizing sequence complementarily. Loss of this snoRNA produces different isoforms of mRNA (Kawaji et al. 2008). This snoRNA is located within the human 15q11q13 locus which has been associated with two human neurological disorders: Prader-Willi Syndrome or Angelman syndrome. These diseases arise from the loss of either paternal, or maternal gene expression within the region, respectively. The loss of the snoRNAs in this region is therefore implicated in the progresses of these diseases, possibly due to inappropriate splicing of the serotonin receptor mRNA (Nicholls et al. 2001).

