piRNAs are a highly conserved class of small RNAs that are likely to have existed since the origin of metazoans (i.e. members of the animal kingdom that are multicellular, heterotrophic eukaryotes that lack cell walls and include sponges all the way up to vertebrates). piRNAs are abundant in most metozoa, but notably absent in plants and fungi. To determine when the piRNA species now observed in most bilataria originally emerged, Grimson, et al used deep sequencing to examine the conservation of Piwi-associated RNAs among various metazoan genomes. This analysis revealed that piRNAs were in fact abundant in both Amphimedon queenslandica (sponge) and Nemtostella vectensis (sea anemone). This is surprising given that mammalian piRNAs are restricted to the germline or soma surrounding germline cells and yet sea sponges and anemones don't have a classical germline - instead they have stem cells that are capable of giving rise to somatic and germline lineages throughout the organism's life. Interestingly, these metazoan species seem to carry both type 1 and type 2 piRNAs, which respectively correspond to pachytene piRNAs and pre-pachytene piRNAs in mammals. Consistent with this notion, the type 2 piRNAs are enriched for transposable elements and appear to have been generated through a ping-pong like mechanism, hence they have a predominant 5' U and a conserved A at position 10. In contrast, the type 1 piRNAs lack these features and comprise a much smaller population of RNAs resembling mammalian pachytene piRNAs. This model of piRNA evolution is further supported by the conservation of at least three Piwi protein homologs and one hen1 gene (i.e. that encodes the methyltransferase required for 2'-0-methylation of piRNAs) in all species from which piRNAs were cloned.
In their paper, Grimson et al make two intriguing observations that 1) a feed-forward amplification loop is required for type 2 piRNA biogenesis 2) this loop generates small RNAs that are then targeted towards the most active transposon species. Taken together, these observations lead the authors to suggest that piRNAs may be one of the main drivers of transposon diversity in animals and are thus likely to play an important role in driving mammalian evolution. Since the time that piRNAs were first cloned and sequenced, it became apparent that most Piwi-associated RNAs derive from finite numbers of clusters around the genome. Though individual piRNA sequences are poorly conserved, these clusters are tightly conserved in syntenic regions on the genome. New work on the evolution of rodent piRNAs suggests that most clusters have arisen through ectopic recombination events whereby long sequences become inserted close to regions with flanking chromosome-specific repeat elements. Intriguingly, the rate of piRNA cluster expansion is much higher than that of any known mammalian gene family and is thus likely to be driven by positive selection, possibly due to an ever-increasing need to silence expanding populations of mammalian transposons (Assis, 2009).
- Piwi proteins: Bind piRNAs and cleave their 5' ends through slicer activity
- Hen-1: a conserved methyltransferase that gives the 2' O-methyl modification on all piRNAs
- Zucchini and Squash: putative 3' endonucleases that determine the 3' end of secondary piRNAs in flies
- Armitage and Spindle-E: RNA helicases required for piRNA generation in flies
- Heterochromatin Protein-1 (HP1a): a chromatin-associated protein that binds PIWI and is likely required for PIWI-induced silencing of transgenes in heterochromatin
- RecQ: In rat it has been demonstrated that the piRNA silencing complex (piRC) interacts with RecQ, an ATP-dependent RNA helicase that shows strong homology to the Neurospora helicase QDE-3 and may be required for piRNA-induced transcript silencing.
Studies of Argonaute family members outside the Piwi clade demonstrate that RNA-Argonaute complexes are capable of cleaving perfectly-matched RNA target sequences or silencing translation of imperfectly paired target mRNAs. Indeed it is likely that pre-pachytene piRNAs, which are enriched in repeat elements, cleave retrotransposon targets through the formation of perfectly matched double-stranded intermediates which arise as part of the ping-pong model. This theory is supported, indirectly, by the observation of decreased transposon activation observed in the absence of Mili or Miwi-2. In mammals, Piwi proteins have been implicated in methylating DNA and thus silencing transcription from those loci. For example, both Mili and Miwi-2 null mutants show demethylation of LINE1 elements and that demethylation is correlated with derepression of those elements (Aravin, 2007; Carmell, 2007). It is still unclear, however, if Piwi's effects on methylation marks is direct or indirect and if it requires the presence of piRNAs. In contrast to Mili and Miwi-2, increased transposon activation has never been observed in Miwi-deficient mice; furthermore it remains obscure what role Miwi-associated pachytene piRNAs play in spermatogenesis or why Miwi-null mice show post-meiotic spermatogenic arrest. Interestingly, Grivna et al observed that Miwi is associated with polysomes throughout spermatogenesis and as polysomes increase in developing spermatocytes, so too does the amount of associated Miwi protein. Interestingly, Miwi was also found to associate with several mRNAs, which it recognizes through the cap binding complex. This would suggest that Miwi-associated piRNAs may be capable of silencing target mRNA transcripts; however, this model is complicated by the fact that Miwi interacts with several miRNAs, which may be mediating translational repression. It remains to be determined what the relationship is between Miwi-associated miRNAs and piRNAs and if these miRNAs can in fact block translation of specific mRNA transcripts by binding the 3' UTR.
In drosophila it has been observed that Piwi associates with transposable elements that are enriched in heterochromatin. Some researchers have demonstrated that Piwi association with heterochromatin is required for Piwi-induced silencing of transposable elements and that this association is likely to occur through Piwi binding the heterochromatin binding protein HP1a (Brower-Toland, 2007). A detailed mechanism of these silencing has yet to be described and it remains unclear if phenomenon exists outside of flies.
Since the discovery of the first Piwi proteins in flies it has become apparent this class of Argonautes is important for mammalian reproduction, though it is still controversial what mechanistic role Piwi-interacting small RNAs play in the Piwi-null fertility phenotype. Mili and Miwi-2 deficient mice show arrested spermatogenesis in early prophase 1, around pachytene stage (Kuramochi-Miyagawa, 2004; Carmell, 2007). In contrast, Miwi-deficient mice show arrested spermatogenesis at the round spermatid stage. The timing of meiotic arrest in the absence of Mili and Miwi-2 suggests that these proteins may facilitate synaptonemal complex formation and crossing over during prophase 1. Interestingly, miwi2 mutants show high levels of DNA break formation, suggesting a defect in DNA repair or synapsis. This observation is similar to Drosophila piRNA pathway mutants, which show germline DNA damage concominant with overexpression of transposable elements and subsequent loss of more mature germ cells through the activation of apoptotic pathways. Given observations that murine Mili and Miwi-2 associate with pre-pachytene piRNAs that are enriched in transposable elements, it's likely that spermatogenic arrest in these mouse mutants is a result of transposon- and retroelement-induced DNA damage. Indeed, mili and miwi-2 mutations in mice are associated with derepression of retroelement trascripts (Aravin, 2007; Carmell, 2007). Interestingly the murine Piwi homologs are not required for female germline maintenance, though this sex-specific difference may reflect the fact that oocytes don't complete prophase 1 until after fertilization and thus a DNA-damage phenotype could be rescued by sperm.
As described above, the founding member of the Piwi gene family was identified in drosophila as a mutation that gives rise to defective oogenesis and loss of germline stem cells (Lin, 1997). As in mice, this mutant phenotype is likely due to the activation of DNA damage checkpoints in the male and female germline. Indeed, mutations in the piRNA pathway lead to significant overexpression of retrotransposons and transposon mobilization in the male fly germline. In contrast, there is no increased transposon mobilization in the female germline of Piwi-deficient flies, so the piRNA pathway may be playing distinct roles in DNA damage repair in this setting.
Somatic piRNAs have recently been discovered in the Drosophila germline and appear to be synthesized by a ping-pong-independent mechanism (Malone, 2009; Li, 2009). Though the exact function of somatic piRNAs is not understood, most of these sequences map to the flamenco locus, which specifically targets long terminal repeat (LTR) elements. In addition, somatic piRNAs appear necessary for female germline maintenance, as ovarian soma undergoes rapid depletion in the absence of Piwi proteins and is no longer able to support germline growth. Taken together, these data suggest that piRNAs may be important for silencing transposable elements in somatic DNA and thus for maintaining the genome integrity of somatic tissues.
Similar to mouse and drosophila Piwi proteins, Zebrafish homologs are required for spermatogenesis and are found to associate with small RNAs that are enriched for repeat elements. Intriguingly, Ziwi-deficient fish are all male, which strongly suggests a yet to be understood role for piRNAs in zebrafish sex determination.
Aravin AA, Hannon GJ and J Brennecke. The Piwi-piRNA pathways provides an adaptive defense in the transposon arms race. 2007. Science 318: 761-4. PMID: 17975059.
Farazi TA, Juanek SA and T Tuschl. The growing catalog of small RNAs and their association with distinct Argonaute/Piwi family members. 2008. Development 135: 1201-14. PMID: 18287206.
Kattenhoff C and W theurkauf. Biogenesis and germline functions of piRNAs. 2007. Development 135: 3-9. PMID: 18032451.
Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Lovino M, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, Chien M, Russo JJ, Ju J, Sheridan R, Sandler C, Zavolan M and T Tuschl. A novel class of small RNAs bind to MILI protein in mouse testes. 2006. Nature 442: 203-7. PMID: 16751777.
Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K and GJ Hannon. Developmentally regulated piRNA cluster implicate MILI in transposon control. 2007. Science 316: 744-747. PMID: 17446352.
Assis R and AS Kondrashov. Rapid repetitive element-mediated expansion of piRNA clusters in mammalian evolution. 2009. PNAS 106: 7079-7082. PMID: 19357307.
Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam T and GJ Hannon. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. 2007. Science 128: 1089-103. PMID: 17346786.
Brennecke J, Malone CD, Aravin AA, Sachidanadam R, Stark A, and GJ Hannon. An epigenetic role for maternally inherited piRNAs in transposon silencing. 2008. Science 322: 1387-92. PMID: 19039138.
Brower-Toland B, Findley SD, Jiang L, Liu L, Yin H, Dus M, Zhou P, Elgin SC and H Lin. Drosophila PIWI associates with chromatin and interactly directly with HP1a. 2007. Genes Dev 21: 2300-11. PMID: 17875665.
Carmell MA, Xuan Z, Zhang MQ and GJ Hannon. The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance and tumorigenesis. 2002. Genes and Development 16: 2733-2742. PMID: 12414724.
Carmell MA, Girard A, van de Kant HJG, Bourc'his D, Bestor TH, de Rooij DG and GJ Hannon. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. 2007. Developmental Cell 12: 503-514. PMID: 17395546.
Deng W and H Lin. miwi, a murine homolg of piwi, encodes a cytoplasmic protein essential for spermatogenesis. 2002. Developmental Cell 2: 819-830. PMID: 12062093.
Girard A, Sachidanandam R, Hannon GJ and MA Carmell. A germline-specific class of small RNAs binds mammalian Piwi proteins. 2006. Nature 442: 199-202. PMID: 16751776.
Grimson A, Srivastava M, Fahey B, Woodcroft BJ, Chiang HR, King N, Degnan BM, Rokhsar DS and DP Bartel. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. 2008. Nature 455: 1193-7. PMID: 18830242.
Grivna ST, Beyret E, Wang Z and H Lin. A novel class of small RNAs in mouse spermatogenic cells. 2006. Genes and Development 20: 1709-1714. PMID: 16766680.
Grivna ST, Pyhtila B and H Lin. MIWI associates with translational machniery and PIWI-interacting RNAs (piRNAs) in regulating spermatogenesis. 2006b. PNAS 103: 13415-20. PMID: 16938833.
Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H and MC Siomi. A slice-mediated mechanism for repeat-associated siRNA 5' end formation in drosophila. 2007. Science 315: 1587-90. PMID: 17322028.
Houwing S, Kamminga LM, Berezikov E, Conembold D, Girard A, van den Elst H, Filippov DV, Blaser H, Raz E, Moens CB, Plasterk RHA, Hannon GJ, Draper BW and RF Ketting. A role of Piwi and piRNAs in germ cell maintenance and transposon silencing in zebrafish. 2007. Cell 129: 69-82. PMID: 17418787.
Kirino Y, Kim N, de Planell-Saguer M, Khandros E, Choriorean S, Klein PS, Rigoutsos I, Jongens TA, and Z Mourelatos. Arginine methylation of Piwi proteins catalysed by dPRMT5 is required for Ago3 and Aub stability. 2009. Nature Cell Biology 11: 652-8. PMID: 19377467.
Kuramochi-Miyagawa S, Tuhru K, Ijiri TW, Isobe T, Asada N, Fujita Y, Ikawa M, Iwai N, Okabe M, Deng W, Lin H, Matsuda Y and T Nakano. Mili, a mammalian member of the piwi family gene, is essential for spermatogenesis. 2004. Development 131: 839-849. PMID:
Li C, Vagin VV, Lee S, Xu J, Ma S, Xi H, Seitz H, Horwich MD, Syrzycka M, Honda BM, Kittler EL, Zapp ML, Klattenhoff C, Schulz N, Theurkauf WE, Weng Z, and PD Zamore. 2009. Collapse of germline piRNAs in the absence of argonaute3 reveals somatic piRNAs in flies. Cell 137: 509-521. PMID: 19395009.
Lin H and AC Spradling. A novel group of pumilio affects that asymmetric division of germline stem cells in the Drosophila ovary. 1997. Development 124: 2463-2476. PMID: 9199372.
Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, and GJ Hannon. Specialized piRNA pathways act in germline and somatic tissues of the drosophila ovary. 2009. Cell 137: 522-535. PMID: 19395010.
Qiao D, Zeeman AM, Deng W, Looijenga LH and H Lin. Molecular characterization of hiwi, a human member of the piwi gene gamily whose overexpression is correlated to seminomas. 2002. Oncogene 21: 3988-99. PMID: 12037681.
Sharma AK, Nelson MC, Brandt JE, Wessman M, Mahmud N, Weller KP and R Hoffman. Human 34(+) stem cells express the hiwi gene, a human orthologue of the Drosophila gene piwi. 2001. Blood 97: 426-34. PMID: 11154219.
Wang G and V Reinke. A C. elegans Piwi, PRG-1, regulates 21U-RNAs during spermatogenesis. 2008. Current Biology 18: 861-7. PMID: 18501605.
Watanabe T, Takeda A, Tsukiyama T, Mise K, Okuno T, Sasaki H, Minami N and H. Imai. Identification of two novel classes of small RNAs in the mouse germline: retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. 2006. Genes and Development 20: 1732-1743. PMID: 16766679.
To date piRNA biogenesis remains incompletely understood. In flies, and possibly mammals, primary piRNAs are likely generated through direct transcription of long single-stranded RNAs, and secondary piRNAs are generated via a "ping-pong mechanism" illustrated above. Primary piRNAs are thought to be transcribed from non-overlapping genomic clusters with strong strand bias or that reveal bidirectional divergent transcription from opposing strands starting from a single site. This model was first proposed based on deep sequencing piRNA from flies (Gunawardane, 2007; Brennecke, 2007). This sequencing revealed that piRNAs bound to Piwi and Aub usually have uridine bases at their 5' ends and that the first 10 nucleotides that bound Piwi or Aub are reverse complements to the first 10 nucleotides that bound Ago3. Furthermore, all three Piwi proteins in flies (Piwi, Aub and Ago3) have active 5' endonuclease domains that are capable of providing the slicer activity that defines the 5' end of the target RNA. It still remains unclear what exonuclease defines the 3' end of the target transcript, though the proteins Zucchini and Squash have been proposed as candidate 3' nucleases in Drosophila. Based on in vitro data from flies and zebrafish it appears that piRNA biogenesis occurs independently of dicer, which is logical given that piRNAs are longer than known dicer products, which are usually 21-22 nt. In flies, and perhaps other species, piRNA generation additionally requires two RNA helicases, Armitage and Spindle-E. Once generated, piRNAs become 2'-O-methylated at their 3' ends by the HEN1 methyltransferase. Though the functional consequence of this modification is unclear, piRNAs generated in HEN1-deficient cells are often uridylated and show increased turnover, suggesting that methylation protects 3' ends from degradation, as is observed in plant miRNAs.
Additionally obscure is how the cyclical ping-pong model first becomes initiated, as this mechanism explains only the expansion of "secondary" piRNA populations. One explanation for how primary piRNAs are synthesized may be explained by the observation that in some species (i.e. zebrafish) piRNAs are only maternally transmitted (Houwing,2007). In flies too it appears that piRNA-mediated transposon control can be inherited only from females (Brennecke, 2008). These observations in flies have been deduced through the study of a process called hybrid dysgenesis, which is a phenomenon whereby crosses between Drosophila strains with different transposons produce sterile progeny. Interestingly, this phenotype is expressed only when the transposable element is paternally transmitted, which lead the authors to hypothesize the existence of a maternally transmitted factor that can silence transposable elements in progeny. Using distinct P- and I-element-induced hybrid dysgenesis models, the authors showed that the piRNAs targeting specific P- or I-elements are all acquired maternally and none are paternally-derived. To date it is unclear if a mechanism such as this could explain the generation of primary piRNAs that feed into the ping-pong model.
To date it remains to be seen how broadly applicable the ping-pong mechanism is to piRNA biogenesis in different species. Some evidence suggests that piRNAs may not be restricted to the germline, as had been thought previsouly, but in fact may be found in somatic ovarian tissue. Interestingly, generation of non-germline piRNAs appears to occur independently of a ping-pong mechanism (Li, 2009; Malone, 2009). In addition, the ping-pong mechanism describes the biogenesis of only prepachytene piRNAs, but is not like to describe how mammalian pachytene piRNAs or C elegans 21U RNAs, which are relatively depleted of repeat elements, become synthesized. It's possible that a conserved 5' sequence upstream of 21 U RNAs in C elegans is required for 5' end processing. Alternatively, the 26 G small RNAs in C elegans could in theory pair with the conserved Uridine in 21U piRNAs, though given that both these bases reside on the 5' end of the small RNAs it's unlikely that they arise from reciprocal cleavage events.
To date little is understood about how piRNA biogenesis is regulated, outside the observation that piRNAs increase or decrease depending on the relative abundance of their respective Piwi binding partners. Thus a better understanding of Piwi regulation may provide important mechanistic insight into the regulation of piRNA biogenesis. Recent work by several groups suggests that all Piwi proteins and their homologues have conserved arginine-rich regions that become methylated and that this methylation is required for Piwi binding to tudor domain-containing proteins (such as Tudor) required for spermatogenesis. In the fly, arginine methylation is required for stabilization of Piwi homologues Ago3 and Aub, and in the absence of the methylene transferase that introduces these modificaitons (pRMT5), these Piwi proteins are rapidly degraded (Kirino, 2009).
Shown below are two representative examples of pre-pachytene and pachytene piRNAs cloned from mouse testis. Based on the the ping-pong model of pre-pachytene piRNA biogenesis we would expect that these small RNAs would have conserved 5'U and A nucleotides at position 10, which is exactly what is observed for mmu_piR_036492. Further examination of the genomic context of this piRNA reveals that it resides within a larger cluster of piRNAs that show substantial overlap with LTR, SINE and LINE elements. In contrast, piR-1, which is a pachytene piRNA, maps to an intergenic region that is relatively depleted of these transposable elements. Also note that piR-1 does not carry a conserved A at position 10, consistent with the observation that this class of piRNAs is synthesized through a ping-pong-independent mechanism. While individual clusters of piRNAs are required to suppress transposable elements, researchers have yet to demonstrate if individual piRNAs are necessary or sufficient for this silencing activity.

(Genomic windows were created using UCSC Genome Browser Mouse Build Feb 2006)
To date there are no descriptions in the literature of using piRNAs as a tool; however, based on knowledge of piRNA biogenesis mechanisms and their probable role in human disease, it's possible to imagine several potential uses of these small RNAs. For example, it is well documented that the human Piwi homolog Hiwi is frequently overexpressed in seminomas (Qiao, 2002). Given preliminary studies in mice, it is likely that piRNAs are also overexpressed in this setting. Thus, it may be possible to use piRNA expression profile signatures to stage seminoma tumors or predict response to specific treatments. In fact, one research group has already filed a patent application to use piRNAs as biomarkers for specific diseases. In the miRNA field, intensive research is currently underway to identify miRNA biomarkers in blood serum, despite little mechanistic evidence for miRNA secretion outside of cells. If researchers are successful in identifying these secreted miRNAs, it may be possible to identify serum-specific piRNAs that are correlated with various disease states. Taking a different approach, it's also possible that human Hiwi may be a good drug target for preventing tumorigenesis. Not only is it overexpressed in testicular seminomas, one study found that it is overexpressed in CD34+ hematopoietic stem cells, and its overexpression (in a leukemia cell line) limited stem cell proliferation by inducing apoptosis (Sharma, 2001).
Another potential tool that may arise from the study of piRNAs is related to the ping-pong biogenesis model. In mammals, piRNAs are a rare form of small RNA that appear capable of amplifying to give secondary RNA species. By comparison, secondary small RNAs are frequently observed in C elegans and are thought to explain the phenomenon of small RNA "spreading" observed when siRNAs are introduced into a local areas of the organism. The secondary small RNAs observed in C elegans, however, arise through RNA-dependent RNA polymerase (RDRP) activity, which is thought to be absent in vertebrates (though is still a point of contention among researchers). The ping-pong biogenesis model provides a potentially useful technique for amplifying small RNAs in the absence of RDRP activity, and may be useful for generating large numbers of small RNAs directed against specific RNA transcripts in vitro or in vivo.
Before it was known that Piwi-interacting RNAs (piRNAs) are associated with argonaute proteins of the Piwi clade, this class of small RNAs was given a variety of different names in the literature. One of the first groups to identify this class of RNAs as unique in mammals (based on their longer size as compared to microRNAs) named them germline small RNAs or gsRNAs, based on their high prevalence in the male germline (Watanabe, 2006), though this nomenclature is used uncommonly. Another nomenclature controversy persists in the field regarding the conservation of piRNAs between different species. For example, in some species piRNAs sequences frequently map to transposable and/or repeat elements and are thus referred to as repeat-associated small interfering RNAs or rasiRNAs, which are now considered a specific type of piRNA. In addition, it is now recognized that the class of 21U RNAs that are highly expressed in the C. elegans germline do in fact interact with conserved Piwi proteins and are now considered bona fide piRNAs (Wang, 2008), despite the fact that they are still referred to as 21U RNAs.
Similar to the miRBase database for miRNAs, all unique cloned piRNAs have been assigned accession numbers and can be found catalogued online at piRNABank. Accession numbers consist of a three letter species designation, followed by the letters “piRNA” and a six digit number that is assigned to sequences roughly in the order that they were cloned and reported in the literature. For example, H. sapiens piRNA-1 is designated hsa_piR_000001.
The first Piwi protein was discovered in 1997 in a Drosophila screen for genes that control asymmetric cell division in the germline (Lin, 1997). In this original study, Lin et al screened a collection of single P-element enhancer-trap female sterile mutants that were shown by immunofluorescence and electron microscopy to undergo asymmetric division in germline stem cells. This screen identified a novel gene that the authors named piwi for P-element induced wimpy testis, which refers to the strong spermatogenesis and sterility defect observed in piwi mutant males.
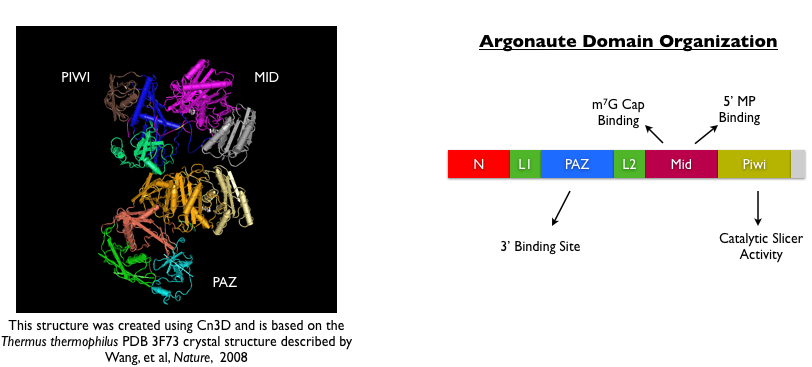 Several years after this initial discovery, three piwi homologues were identified in mammals and were shown to be required in spermatogenesis (Wei, 2002; Kuramochi-Miyagawa, 2003; Carmell, 2007). Detailed genome searches for other Piwi-like genes revealed that this class of proteins actually comprises a larger family of Argonaute proteins that contain conserved PAZ and PIWI domains, shown above (Carmell, 2002). Based on studies that other Argonaute family members were required for miRNA biogenesis, four different research groups simultaneously postulated that germline-specific Piwi proteins may be directing small RNA biogenesis. Indeed, all four groups published papers in 2006 reporting the discovery of a novel class of small RNAs called piwi-interacting or piRNAs that were highly expressed in the male germline. Two of the four groups identified this population of small RNAs simply by running whole testis RNA on a gel and using ethidium bromide to observe a striking population of small RNAs that were unique to testes and notably absent in all somatic tissues (Watanabe, 2006; Grivna, 2006). The other two groups identified small RNAs by immunoprecipitating either the Miwi or Mili proteins (i.e. murine Piwi homologs), running the associated nucleic acid products on a gel and sequencing them (Aravin, 2006; Girard, 2006). Compared to miRNAs, which were the only well-studied population of argonaute-dependent small RNAs in mammalian cells, piRNAs shared some uniquely distinct features: piRNAs were found to be slightly larger then miRNAs (24-30 nt as compared to 21-22 nt), they showed a strong preference for a uridine at the 5’ end, and they mapped to distinct, non-overlapping clusters in the genome that show a strong strand-selection bias. In addition, many piRNA sequences mapped to repeat sequences such as IAP, B1 and LINE elements.
Several years after this initial discovery, three piwi homologues were identified in mammals and were shown to be required in spermatogenesis (Wei, 2002; Kuramochi-Miyagawa, 2003; Carmell, 2007). Detailed genome searches for other Piwi-like genes revealed that this class of proteins actually comprises a larger family of Argonaute proteins that contain conserved PAZ and PIWI domains, shown above (Carmell, 2002). Based on studies that other Argonaute family members were required for miRNA biogenesis, four different research groups simultaneously postulated that germline-specific Piwi proteins may be directing small RNA biogenesis. Indeed, all four groups published papers in 2006 reporting the discovery of a novel class of small RNAs called piwi-interacting or piRNAs that were highly expressed in the male germline. Two of the four groups identified this population of small RNAs simply by running whole testis RNA on a gel and using ethidium bromide to observe a striking population of small RNAs that were unique to testes and notably absent in all somatic tissues (Watanabe, 2006; Grivna, 2006). The other two groups identified small RNAs by immunoprecipitating either the Miwi or Mili proteins (i.e. murine Piwi homologs), running the associated nucleic acid products on a gel and sequencing them (Aravin, 2006; Girard, 2006). Compared to miRNAs, which were the only well-studied population of argonaute-dependent small RNAs in mammalian cells, piRNAs shared some uniquely distinct features: piRNAs were found to be slightly larger then miRNAs (24-30 nt as compared to 21-22 nt), they showed a strong preference for a uridine at the 5’ end, and they mapped to distinct, non-overlapping clusters in the genome that show a strong strand-selection bias. In addition, many piRNA sequences mapped to repeat sequences such as IAP, B1 and LINE elements.
About a year after these initial discoveries, more detailed analyses of piRNA sequences in the germline suggested that they may actually represent two classes of piRNAs of different lengths, one ≈ 30 nt and another several bases shorter. Indeed it was discovered that mammalian piRNAs can be divided into two differentially regulated subclasses referred to as pachytene (29-31 bp) and pre-pachytene (26-28 bp) piRNAs (Aravin, 2007). It's likely that this subdivision of piRNAs was initially overlooked because pachytene piRNAs are tremendeously abundant and thus outcompeted prepachytene piRNAs and miRNAs (by a factor of 10:1) in the original testis small RNA cloning experiments. As the name suggests, pachytene piRNAs first become visible in spermatogonia around the pachytene stage of meiosis (when synapse and crossing over occurs) and begin to decrease when sperm reach the haploid round spermatid stage and spermiogenesis begins. In contrast, pre-pachytene piRNAs are expressed in spermatogonia prior to meiosis and become depleted starting at mid-pachytene. While pachytene piRNAs associate with Miwi, pre-pachytene piRNAs preferentially associate with Mili or Miwi-2. In addition, prepachytene piRNAs are enriched in genome repeat elements while pachytene piRNAs are relatively depleted of repeat elements, suggesting that pre-pachytene piRNAs likely play a role in silencing transposable elements. Researchers have yet to identify a biological role for pachytene piRNAs, and it still remains unknown if the absence of these RNAs is what leads most-meiotic spermatogenesis arrest in Miwi-null mice.
Though researchers have identified no direct role for piRNAs in human disease, the human gene locus homologous to piwi, hiwi, has been implicated in several diseases:
- The hiwi gene locus is linked to the onset of testicular germ cell tumors
- Seminomas, which comprise 40% of all testis tumors and are thought to arise from the germinal epithelium of seminiferous tubules, frequently overexpress Hiwi (Qiao, 2002).
- Loss of the hiwi gene locus has been associated with ambiguous genitalia and testicular atrophy
- Expression of Hiwi is positively correlated with cell proliferation in the setting of pancreatic and gastric cancers


