Given the essential role played by rRNAs in the process of mRNA translation, it is not surprising that rRNAs are highly conserved among species through evolutionary history. In fact, rRNAs are present in all extant species examined to date, and the functionally important rRNA-based peptidyl transferase center, which catalyzes peptide bond formation in the ribosome, has even been recovered through artificial selection from a random sequence pool (Welch et al., 1997). Not surprisingly, rRNA sequences have been widely used in phylogenetic studies. See potential as a tool.
In general, residues located on the surface of the rRNA molecule evolve faster. Additionally, different rRNA secondary structures (such as loops, stems and bulges) evolve at different rates in different lineages (Smit et al., 2007). For instance, in prokaryotes, stems evolve faster than loops; in eukaryotes, however, the reverse is true.
1. Ribosome Database Project website: http://rdp.cme.msu.edu/
2. European Ribosomal RNA Database website: http://bioinformatics.psb.ugent.be/webtools/rRNA/
Composition of the Ribosome
In prokaryotic ribosomes, the large subunit contains over 30 ribosomal proteins apart from the rRNAs, while the small subunit contains about 20 ribosomal proteins besides the 16S rRNA. In eukaryotic ribosomes, the large subunit contains about 50 ribosomal proteins apart from the rRNAs, while the small subunit contains over 30 ribosomal proteins besides the 18S rRNA.
Process of Translation
rRNAs in the ribosome cooperate with tRNAs to translate mRNAs into peptides. This process also requires the cooperation of protein initiation, elongation, release and ribosome recycling factors. See mechanism of action.
Overall Process of Translation
The process of translation can be divided into three distinct phases - initiation, elongation and termination. In prokaryotes, initiation involves the base-pairing of a consensus sequence, known as the Shine-Dalgarno sequence (Shine and Dalgarno, 1975), about 6-10 nucleotides upstream of the mRNA start codon to a complementary sequence near the 3' end of the 16S rRNA in the small ribosomal subunit. This interaction helps to to position the mRNA start codon within the ribosome, and is facilitated by a group of protein initiation factors that also aid in the subsequent recruitment of the large ribosomal subunit. In eukaryotes, on the other hand, initiation involves the binding of protein initiation factors (eIF4E and eIF4G) to the 5' cap of the mRNA. The initiation factors then help to recruit the small ribosomal subunit, bearing a methionine-charged initiator tRNA, to the 5' end of the mRNA. Subsequently, the small ribosomal subunit moves in the 5' to 3' direction down the mRNA, "scanning" for the presence of an AUG start codon, which is present within a consensus sequence known as the Kozak sequence (Kozak, 1987). Once the start codon is located, the initiation factors dissociate, so that the large ribosomal subunit can now bind and complete the ribosome. The methionine-bearing initiator tRNA will then be positioned within the P-site of the ribosome.
During elongation, an aminoacyl-tRNA whose anticodon is complementary to the mRNA codon positioned within the A-site enters the ribosome, bearing a GTP-bound elongation factor known as eEF-1 (EF-Tu in prokaryotes) (reviewed in Steitz, 2008). Upon codon-anticodon base-pairing, three residues of the 16S rRNA in the small subunit alter their conformations to interact with the anticodon and stabilize the tRNA-mRNA complex (Ogle et al., 2001). eEF-1 then hydrolyzes the GTP bound to it, aided by stimulation from an rRNA-based GTPase center in the large ribosomal subunit. Subsequently, eEF-1 dissociates from the aminoacyl-tRNA, which is then fully accommodated in the A-site. eEF-1 serves multiple roles in this process. First, it speeds up elongation, as conformational changes experienced by the ribosome during this process are coupled to the conformational changes that eEF-1 undergoes. Second, eEF-1 improves the accuracy of translation by introducing delays (the time required for GTP hydrolysis and eEF-1 dissociation) into the process of elongation, so that incorect aminoacyl-tRNAs have sufficient time to dissociate from the ribosome before peptide bond formation.
Upon the binding of a correct aminoacyl-tRNA in the A-site, the rRNA-based PTC in the large ribosomal subunit catalyzes the formation of a peptide bond between the incoming amino acid (borne by the aminoacyl-tRNA in the A-site) and the growing peptide chain (borne by the peptidyl-tRNA in the P site). This reaction causes the newly extended peptide chain to be transferred from the tRNA in the P-site to the tRNA in the A-site. A second GTP-bound elongation factor, known as eEF-2 (EF-G in prokaryotes), then binds in or near the A-site, hydrolyzes its bound GTP, and dissociates from the ribosome. This conformational change in eEF-2 helps to accelerate translocation of the peptidyl-tRNA in the A-site and deacylated tRNA in the P-site, along with associated mRNA codons, into the P- and E-sites respectively. Subsequently, a new mRNA codon is exposed in the A-site for interaction with the next aminoacyl-tRNA and the deacylated tRNA leaves the ribosome from the E-site. Recently, the hypusine-containing protein eIF5A, which was initially characterized as a translation initiation factor, has also been shown to have a role in translation elongation and is thought to cooperate with eEF-2 in promoting ribosomal translocation (Saini et al, 2009).
During termination, an mRNA stop codon becomes positioned within the A-site of the ribosome. This is recognized and bound by a protein release factor, which mimics the overall shape and charge distribution of a tRNA molecule. The presence of a release factor in the A-site of the ribosome causes the PTC to catalyze the addition of a water molecule to the peptidyl-tRNA in the P-site. This reaction frees the carboxyl end of the peptide, so that it detaches from the tRNA in the P-site and is released into the cytoplasm. The ribosomal subunits then dissociate with the aid of protein factors (EF-G and a ribosome recycling factor in prokaryotes; the initiation factors eIF3, eIF1, eIF1A and eIF3j in eukaryotes), so that the mRNA and tRNA are released (Hirashima and Kaji, 1973; Pisarev et al., 2007).
Peptide Bond Formation Catalyzed by the Peptidyl Transferase Center
Peptide bond formation in the PTC involves a nucleophilic attack by the α-amino group of the aminoacyl-tRNA on the ester-linked carbonyl carbon of the peptidyl-CCA (Figure 3). In the prokaryotic ribosome, the PTC formed by the 23S rRNA brings the substrates (aminoacyl-tRNA in the A-site and peptidyl-tRNA in the P-site) into close proximity and precisely orients them. Binding of the 3' CCA sequence of the aminoacyl-tRNA to the A-site induces a conformational change in the rRNA of the PTC (Schmeing et al., 2005). This change positions the α-amino group of the aminoacyl-tRNA for its nucleophilic attack, at the same time exposing and reorienting the target carbonyl carbon. Prior to this, the target carbonyl carbon is shielded by the P-site rRNA from nucleophilic attack by water molecules.
The energetically most favorable reaction pathway proceeds through a six-membered transition state (Figure 3) in which proton shuttling occurs via the 2'-OH of the A76 residue of the peptidyl-tRNA (Weinger et al., 2004; Behringer and Rodnina, 2007). This reaction is facilitated by the ordering of water in the active site, and is also modulated by conformational changes at the active site that may be induced by protonation. In addition, it is thought that the ribosome provides a suitable electrostatic environment that stabilizes the highly polar transition state, by shielding the reaction from bulk water and/or aiding the proton shuttle that forms the leaving group.
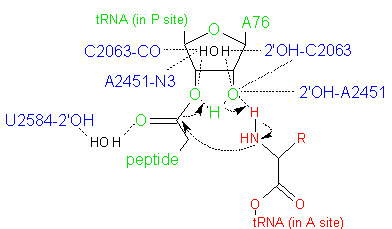
Figure 3. Hypothesized six-membered transition state in the peptidyl transferase reaction, in which proton shuttling occurs via the 2'-OH of the A76 residue of the peptidyl-tRNA. Atoms in red are from the aminoacyl-tRNA; atoms in green are from the peptidyl-tRNA; and atoms in blue are from the PTC active site. Water molecules are shown in black.
Two-thirds of the ribosome are composed of rRNAs while the remaining one third is made up of proteins. Indeed, rRNAs are largely responsible for the 3D structure of the ribosome - they fold into precise, compact structures that fit snugly together to form the interior of the ribosome (Ban et al., 2000). Proteins, on the other hand, are found mainly on the surface and serve to stabilize the overall structure by interacting with RNA domains, often using idiosyncratic extensions that reach into gaps in the RNA core. The structure of the large and small ribosomal subunits are shown in Figures 1 and 2, respectively.
Functionally, the three binding sites for tRNAs in the ribosome (namely, the A-, P- and E-sites) are formed principally by rRNAs (Yusupov et al., 2001). Further, the peptidyl transferase center (PTC) that catalyzes peptide bond formation between the incoming amino acid and the growing peptide chain is also formed by rRNA - the 23S rRNA in the large subunit, in the case of prokaryotes (Ban et al., 2000). Hence, the ribosome qualifies as a ribozyme (RNA enzyme). In addition, the decoding center in the small ribosomal subunit, which positions the mRNA and tRNAs, is entirely constructed of rRNA (Schluenzen et al., 2000). Given the indispensable role played by rRNAs in the structure and functions of the ribosome, rRNAs are critical to the process of protein synthesis.
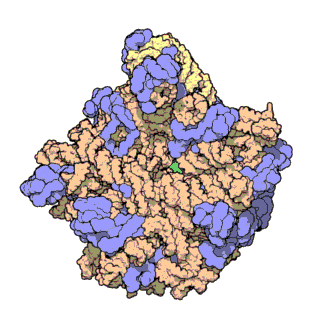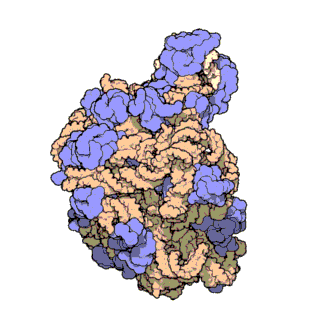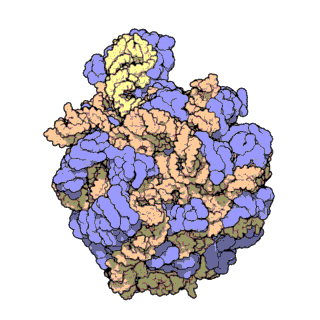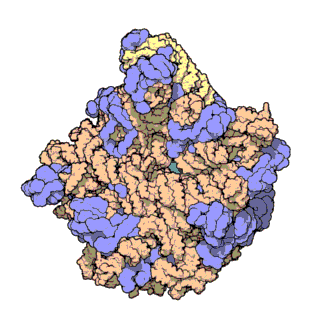
Figure 1. Atomic structure of the large ribosomal subunit from the archaeon Haloarcula marismortui (PDB ID: 1FFK; Ban et al., 2000). Proteins are shown in violet; the two rRNAs are shown in pink and beige; and the active site is depicted by the green patch in the center of the structure.
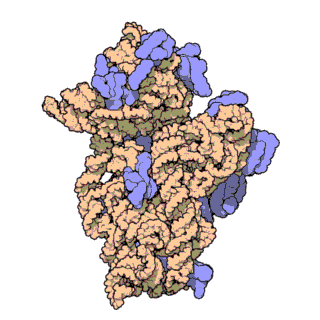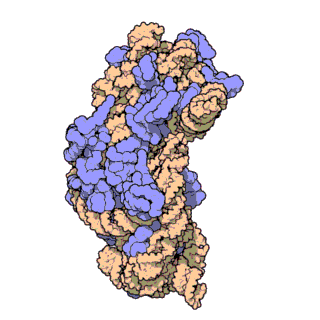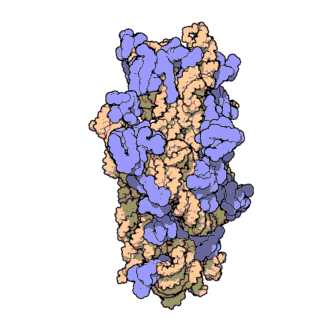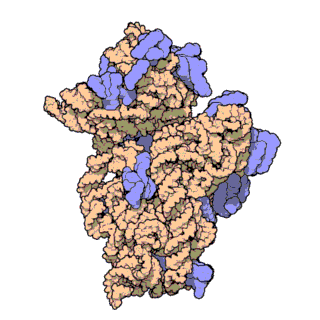
Figure 2. Atomic structure of the small ribosomal subunit from the bacterium Thermus thermophilus (PDB ID: 1FKA; Schluenzen et al., 2000). Proteins are shown in violet and the 16S rRNA is shown in pink.
1. Granneman, S., and Baserga, S.J. (2004). Ribosome biogenesis: of knobs and RNA processing. Exp Cell Res 296, 43-50.
2. Kaczanowska, M., and Ryden-Aulin, M. (2007). Ribosome biogenesis and the translation process in Escherichia coli. Microbiol Mol Biol Rev 71, 477-494.
3. Steitz, T.A. (2008). A structural understanding of the dynamic ribosome machine. Nat Rev Mol Cell Biol 9, 242-253.
Claude, A. (1937). Preparation of an Active Agent from Inactive Tumor Extracts. Science 85, 294-295.
Claude, A. (1938). Concentration and Purification of Chicken Tumor I Agent. Science 87, 467-468.
Claude, A. (1939). Chemical Composition of the Tumor-Producing Fraction of Chicken Tumor I. Science 90, 213-214.
Claude, A. (1940). Particulate Components of Normal and Tumor Cells. Science 91, 77-78.
Palade, G.E. (1955). A small particulate component of the cytoplasm. J Biophys Biochem Cytol 1, 59-68.
Ban, N., Nissen, P., Hansen, J., Moore, P.B., and Steitz, T.A. (2000). The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science 289, 905-920.
Schluenzen, F., Tocilj, A., Zarivach, R., Harms, J., Gluehmann, M., Janell, D., Bashan, A., Bartels, H., Agmon, I., Franceschi, F., et al. (2000). Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution. Cell 102, 615-623.
Yusupov, M.M., Yusupova, G.Z., Baucom, A., Lieberman, K., Earnest, T.N., Cate, J.H., and Noller, H.F. (2001). Crystal structure of the ribosome at 5.5 A resolution. Science 292, 883-896.
Welch, M., Majerfeld, I., and Yarus, M. (1997). 23S rRNA similarity from selection for peptidyl transferase mimicry. Biochemistry 36, 6614-6623.
Kaczanowska, M., and Ryden-Aulin, M. (2007). Ribosome biogenesis and the translation process in Escherichia coli. Microbiol Mol Biol Rev 71, 477-494.
Granneman, S., and Baserga, S.J. (2004). Ribosome biogenesis: of knobs and RNA processing. Exp Cell Res 296, 43-50.
Smit, S., Widmann, J., and Knight, R. (2007). Evolutionary rates vary among rRNA structural elements. Nucleic Acids Res 35, 3339-3354.
Kozak, M. (1987). An analysis of 5'-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res 15, 8125-8148.
Shine, J., and Dalgarno, L. (1975). Determinant of cistron specificity in bacterial ribosomes. Nature 254, 34-38.
Steitz, T.A. (2008). A structural understanding of the dynamic ribosome machine. Nat Rev Mol Cell Biol 9, 242-253.
Ogle, J.M., Brodersen, D.E., Clemons, W.M., Jr., Tarry, M.J., Carter, A.P., and Ramakrishnan, V. (2001). Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292, 897-902.
Saini, P., Eyler, D.E., Green, R., and Dever, T.E. (2009). Hypusine-containing protein eIF5A promotes translation elongation. Nature 459, 118-121.
Hirashima, A., and Kaji, A. (1973). Role of elongation factor G and a protein factor on the release of ribosomes from messenger ribonucleic acid. J Biol Chem 248, 7580-7587.
Pisarev, A.V., Hellen, C.U., and Pestova, T.V. (2007). Recycling of eukaryotic posttermination ribosomal complexes. Cell 131, 286-299.
Schmeing, T.M., Huang, K.S., Strobel, S.A., and Steitz, T.A. (2005b). An induced-fit mechanism to promote peptide bond formation and exclude hydrolysis of peptidyl-tRNA. Nature 438, 520-524.
Weinger, J.S., Parnell, K.M., Dorner, S., Green, R., and Strobel, S.A. (2004). Substrate-assisted catalysis of peptide bond formation by the ribosome. Nat Struct Mol Biol 11, 1101-1106.
Beringer, M., and Rodnina, M.V. (2007). The ribosomal peptidyl transferase. Mol Cell 26, 311-321.
Stock, D.W., and Whitt, G.S. (1992). Evidence from 18S ribosomal RNA sequences that lampreys and hagfishes form a natural group. Science 257, 787-789.
Mallatt, J., and Sullivan, J. (1998). 28S and 18S rDNA sequences support the monophyly of lampreys and hagfishes. Mol Biol Evol 15, 1706-1718.
Cole, J.R., Wang, Q., Cardenas, E., Fish, J., Chai, B., Farris, R.J., Kulam-Syed-Mohideen, A.S., McGarrell, D.M., Marsh, T., Garrity, G.M., et al. (2009). The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37, D141-145.
Wuyts, J., Perriere, G., and Van De Peer, Y. (2004). The European ribosomal RNA database. Nucleic Acids Res 32, D101-103.
Prezant, T.R., Agapian, J.V., Bohlman, M.C., Bu, X., Oztas, S., Qiu, W.Q., Arnos, K.S., Cortopassi, G.A., Jaber, L., Rotter, J.I., et al. (1993). Mitochondrial ribosomal RNA mutation associated with both antibiotic-induced and non-syndromic deafness. Nat Genet 4, 289-294.
Coulbault, L., Deslandes, B., Herlicoviez, D., Read, M.H., Leporrier, N., Schaeffer, S., Mouadil, A., Lombes, A., Chapon, F., Jauzac, P., et al. (2007). A novel mutation 3090 G>A of the mitochondrial 16S ribosomal RNA associated with myopathy. Biochem Biophys Res Commun 362, 601-605.
Yoon, A., Peng, G., Brandenburger, Y., Zollo, O., Xu, W., Rego, E., and Ruggero, D. (2006). Impaired control of IRES-mediated translation in X-linked dyskeratosis congenita. Science 312, 902-906.
In Prokaryotes
The 5S, 16S and 23S rRNAs are synthesized as one primary transcript (reviewed in Kaczanowska et al., 2007). Maturation of the transcript begins before transcription is complete, with rapid formation of secondary structures and binding of ribosomal proteins as soon as binding sites emerge from the polymerase. A the same time, the precursor rRNA undergoes numerous chemical modifications (for instance, conversion of uridines into pseudouridines and addition of methyl, amino, carbonyl or thio groups) and is processed by several RNases (such as RNase III) to generate the mature rRNAs.
rRNA biogenesis in prokaryotes is tightly regulated. When there is a reduction in available nutrients and a consequent fall in the level of aminoacyl-tRNAs, the so-called stringent response is set off, leading to an increase in the level of two small nucleotides, pppGpp and ppGpp, which act as regulators of the stringent response. pppGpp and ppGpp are synthesized from GTP and GDP respectively, using ATP as a phosphate donor. (p)ppGpp binds and inhibits RNA polymerase. As a result, rRNA transcription level is reduced, so that ribosomal proteins in the cytoplasm are now in excess. The surplus ribosomal proteins then bind and inhibit their own mRNAs.
In Eukaryotes
The genes encoding rRNA have an important role in forming the nucleolus. In humans, the rRNA genes encoding a common precursor for the 5.8S, 18S and 28S rRNAs are organized into 10 clusters that occur near the tips of Chromosomes 13, 14, 15, 21 and 22 (the 5S rRNA is encoded by a separate cluster of genes and is transcribed by RNA Polymerase III). During mitosis, the nucleolus disappears as the chromosomes decondense; after mitosis, however, the tips of the 10 chromosomes coalesce to reform the nucleolus.
rRNA biogenesis begins in the nucleolus (reviewed in Granneman and Baserga, 2004). Here, the rRNA genes are transcribed by RNA Polymerase I into a 13,000 bp precursor rRNA, known as the 45S precursor rRNA. First, this precursor undergoes extensive chemical modifications - principally methylations at the 2'-OH position of the nucleotide ribose rings and isomerizations of uridine residues into pseudouridines. These modifications are made at specific positions of the 45S precursor rRNA with the help of several hundred "guide RNAs," which belong to the class of RNAs known as small nucleolar RNAs (snoRNAs). The guide RNAs associate with proteins to form complexes known as snoRNPs, and base-pair with the 45S precursor rRNA to bring RNA-modifying enzymes to specific positions within the precursor rRNA. The modified precursor rRNA then assembles with trans-acting factors and ribosomal proteins, mostly of the small subunit, into a 90S pre-ribosome. Subsequently, cleavage of the 90S pre-ribosome between the sequences of the 5.8S and 18S rRNAs leads to the formation of separate pre-40S and pre-60S ribosomal subunit precursors. These are then exported independently from the nucleus and undergo futher maturation in the cytoplasm to generate the complete ribosome.
Prokaryotes have 3 varieties of rRNA: 5S, 16S and 23S rRNAs. The 5S and 23S rRNAs are found within the large ribosomal subunit, while the 16S rRNA is located in the small ribosomal subunit. Eukaryotes, on the other hand, have at least 4 flavors of rRNA: 5S, 5.8S, 18S and 28S rRNAs. The 5S, 5.8S and 28S rRNAs are found within the large ribosomal subunit, while the 18S rRNA is located in the small ribosomal subunit. Apart from these, the mitochondria and chloroplasts in eukaryotes also have their own rRNAs (for instance, the 12S and 16S mitochondrial rRNAs in mammals), which are grossly similar to those in the prokaryotes, thus lending support to the Endosymbiont Theory.
Because rRNA is highly conserved across species through evolutionary history and is presumably present in the earliest forms of life, genes encoding rRNAs are often sequenced to determine an organism's taxonomic group, infer phylogeny (evolutionary relationships among taxonomic groups) and estimate rates of species divergence. For instance, phylogenetic studies comparing the sequences of the genes encoding 18S and 28S rRNAs in lampreys, hagfishes and gnathostomes (jawed vertebrates) indicate that lampreys and hagfishes form a natural (monophyletic) group separate from the gnathostomes (Stock et al., 1992; Mallatt et al., 1998); these results challenge the once commonly held view, arising from phenotypic analyses, that the lampreys are more closely related to the gnathostomes than to the hagfishes, which form a separate basal lineage.
To date, thousands of rRNA sequences from various species are known and stored in databases such as the Ribosome Database Project (RDP-II) (Cole et al., 2009) and the European Ribosomal RNA Database (Wuyts et al., 2004), which provide researchers with valuable alignment and analysis tools for phylogenetic studies.
In vitro Translation Systems
In vitro, cell-free translation systems are preferred to in vivo gene expression and protein production in situations where the overexpressed protein is toxic to the cell, where the protein is rapidly degraded by endogenous proteases, or where the protein is insoluble. The role played by rRNAs in in vivo translation has been exploited for use in these in vitro translation systems. Both prokaryotic and eukaryotic in vitro translation systems are available - the former typically involves ribosomal preparations from E. coli while the latter involves ribosomal preparations from rabbit reticulocytes or wheat germ. Other components required for these translation systems to work include tRNAs, amino acids, energy sources (ATP, GTP) and an energy-regenerating system (phosphoenol pyruvate and pyruvate kinase for the prokaryotic system, and creatine phosphate and creatine phosphate and creatine phosphokinase for the eukaryotic system). In vitro translation, which uses RNA as a template, has also been coupled to in vitro transcription in some transcription:translation systems that use DNA as a template.
Ribosomal RNA is also known as rRNA. The various species of rRNAs are named based on their sedimentation rates in a centrifuge, measured in Svedberg units (S); the higher the sedimentation coefficient in Svedberg units, the faster the sedimentation rate. For instance, 23S rRNA (found within the the large ribosomal subunit in prokaryotes) refers to an rRNA species with a sedimentation rate of 23S. Larger rRNAs tend to sediment faster, hence 23S rRNA is larger than 16S rRNA.
rRNA as Part of the Microsome
rRNA was first discovered not as a component of the ribosome but as a part of the microsome, which is a vesicle-like artifact that forms from the endoplasmic reticulum and associated ribosomes when cells are lysed. In the late 1930s, Albert Claude was trying to purify the tumor-causing fraction from a form of chicken tumor induced by an RNA virus and found, as expected, that the fraction had a high RNA content (Claude, 1937, 1938, 1939). Unexpectedly, however, he also found that biochemically similar, RNA-rich fractions could be isolated from a variety of other chicken tissues, both embryonic and adult, as well as normal and tumorous (Claude, 1940). Claude concluded (erroneously) that the fractions contained a new, common cell component and coined the term "microsome" (small body) for it. The microsome is distinguished by its high RNA content and later work also indicated that the microsome has a remarkable ability to incorporate labeled amino acids into proteins both in vivo and in vitro. According to modern interpretation, this latter property arises because the microsomal fraction contains the site of protein synthesis in cells.
rRNA as Part of the Ribosome
It was not until the mid-1950s, after significant advancements in electron microscopy techniques, that rRNA was finally recognized as a component of the ribosome in cells. Then, George Palade characterized the ribosome as a new cytoplasmic component that normally assumes the form of a small, round body about 100 to 150 A in diameter (Palade, 1955). He observed that the small granules can be freely scattered in the cytoplasm or found in close association with the cytoplasmic surface of the endoplasmic reticulum. Crucially, he also observed that cytoplasmic basophilia (tendency of the cytoplasm to stain with basic dyes), which is a property that is known to be largely dependent on the presence of RNA, is most closely correlated with the small granules. Hence, Palade concluded that the small granules (i.e. ribosomes) contain a large quantity of RNA (i.e.rRNA).
Crystal Structure
With the advent of advanced X-ray crystallographic techniques, high-resolution atomic structures of the prokaryotic large and small ribosomal subunits were finally obtained in 2000. First, the structure of the large ribosomal subunit from the archaeon Haloarcula marismortui, at 2.4 A resolution, was published (Figure 1; Ban et al., 2000). This study showed that the 3D structure of the large subunit was largely dependent on the rRNAs. Soon after, the structure of the small ribosomal subunit from the bacterium Thermus thermophilus, at 3.3 A resolution, was also published (Figure 2; Schluenzen et al., 2000). This study showed that the decoding center of the small subunit, which positions the mRNA and tRNAs, is wholly composed of rRNA. The following year, the complete structure of the ribosome from Thermus thermophilus, containing bound mRNA and tRNAs at 5.5 A resolution, was published (Yusupov et al., 2000).
Mutations in Mitochondrial rRNA
The main complication of aminoglycoside antibiotics, such as streptomycin, kanamycin and gentamicin, is irreversible hearing loss (or ototoxicity). The natural target of these antibiotics is the bacterial ribosome, in which aminoglycosides bind to rRNA and appear to stabilize mismatched amino acyl-tRNAs, thus leading to misreading of mRNAs during translation. Because susceptibility to aminoglycoside ototoxicity also shows evidence of maternal transmission, it was hypothesized that mutations in mitochondrial rRNA genes may be associated with susceptibility to aminoglycoside ototoxicity (Prezant et al., 1993). By performing linkage studies, the authors found an A1555G point mutation in the mitochondrial 12S rRNA gene that cosegregates with susceptibility to aminoglycoside ototoxicity. In bacteria, the analogous ribosomal nucleotide position is implicated in aminoglycoside binding. The authors suggest that the A1555G substitution in mitochondrial 12S rRNA exerts its effect by increasing aminoglycoside binding, which in turn leads to an increased susceptibility to the detrimental effects of aminoglycosides on translational fidelity. In addition to mutations in the 12S rRNA, a G3090A substitution in the mitchondrial 16S rRNA has been reported to be responsible for the development of myopathy in a young female patient (Coubault et al., 2007).
Changes to rRNA in Cytoplasmic Ribosomes
Apart from mutations in mitochondrial rRNAs, changes to rRNAs in the cytosol have also been implicated in disease. For instance, X-linked dyskeratosis congenita (X-DC) is a progressive congenital disease that leads to skin abnormalities, bone marrow failure and increased risk of cancer. It arises from a mutation in the gene DKC1, which encodes the pseudouridine synthase dyskerin. X-DC patient cell lines exhibit reduced rRNA pseudouridylation, and the mutated ribosomes have been reported to be defective in translating mRNAs with an internal ribosome entry site (IRES) element (Yoon et al., 2006). IRES elements, present within a subset of mRNAs, are structured RNA motifs that directly bind the ribosome during translation initiation, thereby bypassing the need for some of the cap-binding initiation factors. Using an X-DC mouse model and cells from X-DC patients, Yoon et al. showed that the mutated ribosomes are impaired in their ability to translate mRNAs encoding the tumor suppressor p27 and the anti-apoptotic factors Bcl-xL and XIAP, which all harbor an IRES element. The authors further suggest that the defective p27 translation may account for increased tumor susceptibility, while the reduction in Bcl-xL and XIAP levels may result in increased apoptosis of hematopoietic progenitors and stem cells, thus leading to bone marrow failure in X-DC patients.

