rasiRNAs have been identified in plants, Trypanosoma brucei, fission yeast Schizosaccharomyces pombe, Drosophila melanogaster and zebrafish Danio rerio. In mammals, rasiRNAs have not been specifically described, but RNAs that are 22-24 nt in length have been reported in the mouse male germline and they associate with murine homolog of piwi which is part of the Piwi Argonaute subfamily. These RNAs have been more generically termed as piRNAs, because even though they share common features with rasiRNAs, they lack sequences sequences matching repetitive elements. (Watanabe, 2006 and Grivna, 2006)
Like miRNAs and siRNAs, rasiRNAs have a strong preference for pyrimidine residues, especially uridine, at their 5’-most position. However, this is only restricted to the antisense rasiRNA strands. Sense rasiRNAs show no bias at the 5’ ends, though they have a strong preference for adenine at nucleotide 10. Also, rasiRNAs have a monophosphate group at the 5’ end, similar to miRNAs and siRNAs. However, unlike miRNAs and siRNAs which have 2’ and 3’ hydroxyl termini, rasiRNAs have a modification at one of the 2 hydroxyl termini.
In yeast, Dicer is involved in the production of mature rasiRNAs, and the RNAs then associate with Ago1, Chp1 and Tas3 that make up the RITS complex.
In plants, the biogenesis of rasiRNA requires the RNA-dependent RNA polymerases RDR2 and SDE4, the plant homologs dicer-like DCL2 and DCL3 and AGO4.
In flies, rasiRNAs associate with members of the Piwi subfamily of Argonaute proteins. Antisense rasiRNAs associate with Piwi and Aubergine, while sense rasiRNAs associate with Ago3. methyltransferase Pimet binds to the 3’ ends of rasiRNAs to mediate the 2’-O-methylation at the ends. Putative nucleases phospholipase D nuclease Zucchini (zuc) and the RNase HII-related protein Squash (squ) have been shown to physically interact with Aub and is speculated to mediate the 3’ end cleavage of rasiRNAs to produce the mature RNA strands during biogenesis. Putative helicases Spindle-E (Spn-E) and Armitage (Armi) have been implicated in the mechanism of rasiRNA-dependent silencing of chromatin, genomic repeats and retrotransposons, hence they likely bind to the rasiRNA and target RNA sequences (Farazi, 2008)
In yeast, Dcr (yeast Dicer) cleaves long dsRNA derived from repetitive sequences transcribed in both sense and antisense directions to produce rasiRNAs. These rasiRNAs then serve as the guide transcripts in RNA-induced initiation of the transcriptional silencing complex (RITS). The complex is comprised of proteins Ago1, Chp1 (a chromatin binding and histone methylase) and Tas3 (a histone methylase). This complex never leaves the nucleus and binds to DNA where it induces histone and DNA methylation. This leads to the process of transcriptional gene silencing (TGS), and establishes the heterochromatin (Farazi, 2008).
In plants, rasiRNA gene silencing is similar to that in yeast. However, in plants, rasiRNA association with RITS complex is also able to repress repetitive transposable elements. rasiRNAs function through Ago4-dependent mechanisms to direct chromatin modifications, including inducing systemic silencing, mediating histone H3 lysine-9 methylation and using non-CpG asymmetric DNA methylation (Farazi, 2008).
In Drosophila melanogaster, it is speculated that rasiRNAs associate with RNAi silencing components of the piwi Argonaute family and RNA helicases spindle-E and Armitage and induces histone H3 Lys9 methylation and HP1 (a chromatin binding protein) associations in heterochromatin regions, as mutations in the protein complex components result in defects in methylation and HP1 heterochromatin associations (Farazi, 2008).
rasiRNAs have been described in fission yeast, plants, fly and zebrafish, and they generally serve to regulate these processes to ensure genomic stability: (i) guide transcriptional and post-transcriptional gene silencing of repetitive sequence elements, (ii) silence and suppress mobilization of retrotransposons and other selfish genetic transposable elements and (iii) guide chromatin silencing to establish and maintain heterochromatin.
In the fly and zebrafish, rasiRNAs mainly functions in the germline and early embryo development. In the rasiRNA profile of Drosophila melanogaster development, Aravin et al. reported that rasiRNA content was high in the testes and the early embryo, and significantly decreases during development in the transition to adults, in which rasiRNAs only represent about 10% of all small RNAs present. This lends evidence that the downregulation of transposable elements and the establishment of chromatin structure are initiated well early in the germline and during early embryonic development.
In particular, rasiRNAs are important in the Drosophila germline in additional processes, including: (i) maintaining germ cell self-renewal, (ii) establishing axis polarity of the embryo, (iii) regulating pole cell formation and oocyte maturation and (iv) maintaining telomeres.
- Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009 Feb;10(2):94-108. Pubmed.
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009 Feb;10(2):126-39. Pubmed.
- Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell. 2009 Feb 20;136(4):656-68. Pubmed.
- Aravin AA, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, Snyder B, Gaasterland T, Meyer J, Tuschl T. The small RNA profile during Drosophila melanogaster development. Dev Cell. 2003 Aug;5(2):337-50. Pubmed.
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007 Mar 23;128(6):1089-103. Pubmed.
- Brennecke J, Malone CD, Aravin AA, Sachidanandam R, Stark A, Hannon GJ. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science. 2008 Nov 28;322(5906):1387-92. Pubmed.
- Chen X. A marked end. Nat Struct Mol Biol. 2007 Apr;14(4):259-60. Pubmed.
- Djikeng A, Shi H, Tschudi C, Ullu E. RNA interference in Trypanosoma brucei: cloning of small interfering RNAs provides evidence for retroposon-derived 24-26-nucleotide RNAs. RNA. 2001 Nov;7(11):1522-30. Pubmed.
- Farazi TA, Juranek SA, Tuschl T. The growing catalog of small RNAs and their association with distinct Argonaute/Piwi family members. Development. 2008 Apr;135(7):1201-14. Pubmed.
- Grivna ST, Beyret E, Wang Z, Lin H. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 2006 Jul 1;20(13):1709-14. Pubmed.
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC. A slicer-mediated mechanism for repeat-associated siRNA 5' end formation in Drosophila. Science. 2007 Mar 16;315(5818):1587-90. Pubmed.
- Hartig JV, Tomari Y, Förstemann K. piRNAs--the ancient hunters of genome invaders. Genes Dev. 2007 Jul 15;21(14):1707-13. Pubmed.
- Hamilton A, Voinnet O, Chappell L, Baulcombe D. Two classes of short interfering RNA in RNA silencing. EMBO J. 2002 Sep 2;21(17):4671-9. Pubmed.
- Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, van den Elst H, Filippov DV, Blaser H, Raz E, Moens CB, Plasterk RH, Hannon GJ, Draper BW, Ketting RF. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell. 2007 Apr 6;129(1):69-82. Pubmed.
- Saito K, Nishida KM, Mori T, Kawamura Y, Miyoshi K, Nagami T, Siomi H, Siomi MC. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 2006 Aug 15;20(16):2214-22. Pubmed.
- Saito K, Sakaguchi Y, Suzuki T, Suzuki T, Siomi H, Siomi MC. Pimet, the Drosophila homolog of HEN1, mediates 2'-O-methylation of Piwi- interacting RNAs at their 3' ends. Genes Dev. 2007 Jul 1;21(13):1603-8. Pubmed.
- Shpiz S, Kwon D, Rozovsky Y, Kalmykova A. rasiRNA pathway controls antisense expression of Drosophila telomeric retrotransposons in the nucleus. Nucleic Acids Res. 2009 Jan;37(1):268-78. Pubmed.
- Okamoto H, Hirochika H. Silencing of transposable elements in plants. Trends Plant Sci. 2001 Nov;6(11):527-34. Pubmed.
- Pane A, Wehr K, Schüpbach T. zucchini and squash encode two putative nucleases required for rasiRNA production in the Drosophila germline. Dev Cell. 2007 Jun;12(6):851-62. Pubmed.
- Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006 Jul 21;313(5785):320-4. Pubmed.
- Watanabe T, Takeda A, Tsukiyama T, Mise K, Okuno T, Sasaki H, Minami N, Imai H. Identification and characterization of two novel classes of small RNAs in the mouse germline: retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes Dev. 2006 Jul 1;20(13):1732-43. Pubmed.
Most of the current information about the biogenesis of rasiRNAs is based on studies conducted in Drosophila melanogaster. In the fly, there are 5 distinct members of the Argonaute family. These can be classified into 2 different subfamilies, the Ago and Piwi subfamily. The Ago subfamily consists of Ago1 and Ago2 which associate with miRNA and siRNA respectively. Aub, Piwi and Ago3 belong to the Piwi subfamily and are enriched in germline cells and the early embryo. This corresponds to the developmental stage and location exhibiting a rich abundance of rasiRNAs.
It was initially believed that rasiRNAs were derived from double-stranded precursors and processed by ribonuclease type III Dicer, similar to miRNA and siRNAs. This was due to the strong preference for uridine residues at their 5’-most position, as well as the presence of rasiRNAs sequences of both sense and antisense polarity (Aravin, 2003).
However, it was later discovered by Vagin et al. and Saito et al. that rasiRNAs have a distinct and separate mechanism of biogenesis from miRNAs and siRNAs. The authors showed that rasiRNAs likely arise from the antisense strand as a single-stranded precursor, are produced in a Dicer-independent manner and are bound by the Piwi subfamily of Argonaute proteins. Mutations in Piwi subfamily proteins and proteins associated with the rasiRNA mechanism resulted in loss of antisense, but not sense rasiRNA species and disrupted relevant gene silencing, while mutations in Ago subfamily protein/Dicer family proteins did not affect the process. This is in contrast to the production of miRNAs and siRNAs which operate through Dicer via a double-stranded precursor stage and associate with the Ago Argonaute subfamily proteins (Vagin, 2006). The evidence from Vagin et al. also demonstrated that rasiRNAs physically associate with both Piwi and Aub, and hence this suggests that piwi and aub could partially redundant and function in the same pathway to silence selfish genetic elements. They also noted that rasiRNA biogenesis required putative helicases Spindle-E (Spn-E) and Armitage (Armi).
Gunawardane et al. and Brennecke et al in 2007 both reported that Ago3 also associates with rasiRNAs but these RNAs are derived from the sense strand and showed a strong preference for adenine at nucleotide 10. Comparisons of Ago3-associated sense rasiRNAs and Aub-associated antisense rasiRNAs showed complementarities in their first 10 nucleotides. However, it was surprising that similar complementarities were not found between ago3- and piwi-associated rasiRNAs. It may be partially because Piwi has been described to localize to the nucleus, while Ago3 and Aub are cytoplasmic. The authors speculated that piwi-associated rasiRNAs may be functional only at an earlier time during germline development and involved in nuclear RNA silencing. The timepoint tested by the authors in the ovary reflected only rasiRNA biogenesis that operated in the cytoplasm.
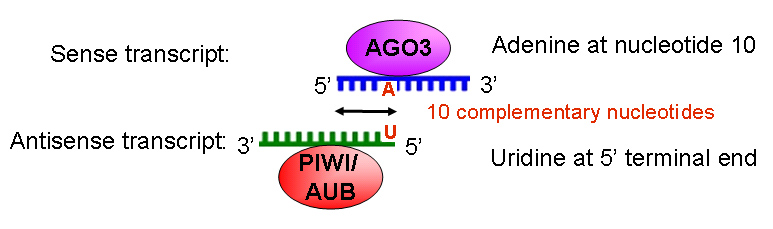
Figure 2. Sense and antisense rasiRNA transcripts. Antisense rasiRNAs have a strong preference for uridine residues at their 5’-most position, while sense rasiRNAs have a strong preference for adenine at nucleotide 10.
The authors proposed a model of rasiRNA 5’ processing called the ping-pong model. In the model, an existing rasiRNA associates with a target RNA of the opposite polarity and the Piwi proteins Piwi/Aubergine or Ago3 depending on the polarity of the RNA strand. The primary rasiRNA template helps to guide the Piwi protein to cleave the RNA transcript and produce the 5’ end of the new rasiRNA and lead to the production of a 2nd rasiRNA. Some Argonaute proteins have been reported to exhibit Slicer activity that cleaves its cognate mRNA target across from nucleotides 10 and 11 as measured from the 5’ end of the small RNA guide strand. Thus, it is likely that Ago3-associated sense rasiRNA can target the antisense RNA molecule via Watson-Crick base pairing and cleaves the Aub- and Piwi-associated rasiRNAs with U at the 5’ ends. Likewise, Aub- or Piwi-associated rasiRNAs slice its cognate RNA target to result in A at nucleotide 10 on the cleaved RNA. This results in an amplification loop to continuously produce additional rasiRNAs in the ovary and testis as well as the early embryo that target transposons. (Gunawardane, 2007 and Chen, 2007)

Figure 3. Current 'ping pong' model of rasiRNA biogenesis.
To date, an unresolved piece of the rasiRNA biogenesis pathway is how the 5’-sliced rasiRNA precursors are defined at their 3' ends and cleaved to result in the mature rasiRNA. This can be acted by an unidentified endonuclease or nibbled by exonuclease and can occur either before or after loading of the resulting cleavage products onto another member of the Piwi subfamily in the rasiRNA production cycle.
However, we do know is that the terminal ribose at the 3’ ends of rasiRNAs is modified. This 3’ terminal modification was inferred by Vagin and Gunawardane when they observed that Drosophila rasiRNAs are resistant to sodium periodate oxidation treatment followed by ß-elimination that would usually result in an RNA shortening by one nucleotide. Saito et al. followed up and demonstrated that the rasiRNAs are 2’-O-methylated at their 3’ ends and this process is mediated by a methyltransferase Pimet, the Drosophila homolog of Arabidopsis HEN1 methyltransferase for microRNAs. A potential function of the 3’ terminal methylation may be to mark the 3’ ends of rasiRNA and protect it from degradation by enzymes (Chen, 2007).
Recently, two proteins containing domains with homologies to nucleases have been implicated in the biogenesis of the rasiRNA 3' end: the Phospholipase D nuclease Zucchini (zuc) and the RNase HII-related protein Squash (squ) (Pane, 2007). Pane et al showed that Zuc and squ physically interacts with Aub piwi protein that is required for the the rasiRNA mechanism in silencing selfish genetic elements. Mutations in these genes abolish the production of rasiRNAs and upregulated the expression of transposable elements and tandem repeats in the Drosophila ovaries and testes and early during oogenesis. Also, telomeres in Drosophila are maintained by the transposition of retrotransposons such as HeT-A and Tart to the chromosome ends and the authors found that zuc and squ mutants also had upregulated levels of these elements leading to telomere elongation and chromosomal abnormalities. The role of rasiRNAs in gene silencing was demonstrated as the zuc and squ mutant failed to suppress Oskar (osk) expression which is one of the genes essential for establishing the axis polarity in the germline. Mutant females are sterile and show dorsoventral patterning defects during oogenesis.
It also remains unclear how the ping-pong biogenesis mechanism is initiated. In the fly, aub and possibly piwi are accumulated at the posterior pole in the oocytes and remains in the polar granules in the early embryos. It is then deposited in the pole cells which are the progenitor germ cells. Hence, this suggests that rasiRNAs which are associated with Piwi proteins can be transmitted via the germline for the next generation, acting as ‘primary seeds’ to launch the rasiRNA biogenesis cycle in the embryos. The maternal loading of Piwi proteins into embryos was also observed in fish. A recent study by Brennecke et al. demonstrated the importance of maternal germline transmission of rasiRNAs in providing transposon resistance. The authors showed that rasiRNAs that are maternally inherited to embryos have an epigenetic regulatory role in transposon silencing and maintains fertility of the embryos. However, if the particular transposon is paternally inherited, this leads to loss of rasiRNA inheritance and sterile progeny in a phenomenon called hybrid dysgenesis (Brennecke, 2008).
In plants, the biogenesis of rasiRNA requires the RNA-dependent RNA polymerases RDR2 and SDE4, the plant DCL3 (Dicer-like) and the plant AGO4.
rasiRNAs are present in both the sense and antisense orientation of all known repetitive sequence elements, such as long terminal repeat (LTR) and non-LTR retrotransposons, DNA transposons, satellite and microsatellite DNA sequences, complex repeats like the Su(Ste) locus, as well as vaguely characterized repetitive sequence motifs (Aravin, 2003). These have been described by several reports which generated libraries of short/small RNAs or conducted northern analyzes.
To date, several RNA profiling studies have been conducted particularly in the fly to search for rasiRNAs that associate with Piwi subfamily proteins – ago3, aub and piwi. For each separate protein, at least a few hundred rasiRNAs have been identified to associate with it.

Table 1. Identified rasiRNAs that associate with each of the piwi subfamily proteins in the fly ovary. The rasiRNAs identified are classified into sense or antisense orientation.
Below are a few representative examples of the repetitive sequence elements:
- LTR retrotransposons – roo, mdg1, gypsy
- Non-LTR retrotransposons – I-element, HeT-A and Tart telomere components
- Complex repeats – Su (Ste) repeats
- Repetitive locus – mst40
- Other heterochromatic repeats – satellite and microsatellite DNA sequences
An example of the rasiRNA cluster locus is the flamenco/COM locus. It is located on the X chromosome and has been shown to regulate the activity of retrotransposons gypsy, Idefix and ZAM. rasiRNAs originating from the flamenco cluster locus are proposed to control the activities of gypsy, Idefix and ZAM in the Drosophila germline.
Some interesting examples:
1. Complex repeats acting in trans: Suppressor of Stellate [Su(Ste)] repeats are located on the Y chromosome and its deletion will derepress Stellate which is located on the X chromosome and result in meiotic abnormalities and male infertility due to crystallization of overexpressed Stellate protein in sperm cells. 25-27nt siRNAs (rasiRNAs) are derived from the strands of Su(Ste) and they are associated with Stellate silencing. Mutants involved in the silencing mechanism results in defects in the female germline and accumulation of transcripts from retrotransposons in the germline (Aravin, 2003).
2. Retrotransposons in telomere elongation: Drosophila does not have telomerases that add short RNA repeats to chromosome ends like eukaryotes. Instead, telomeres are maintained by transpositions of specialized telomeric retrotransposons, namely HeT-A, TART and TAHRE. rasiRNAs derived from the transcription of these elements regulate the silencing of the retrotransposons (Shpiz, 2009).
rasiRNAs are important in regulating gene silencing of repetitive sequence elements and to silence and suppress mobilization of transposable elements. In the genomes of many eukaryotes, including humans, the course of evolution has resulted in a lot of ‘junk DNA’ involving duplications and repeats. rasiRNAs can potentially be engineered to act on transcriptionally active repeated elements to control their expression levels, especially those implicated in diseases. Engineered rasiRNAs can similarly be targeted to retrotransposon elements or to act on heterochromatin DNA in a temporal and spatially regulated manner to aid in the maintenance of heterochromatin and ensure proper silencing and activation of genes. For example, the use of rasiRNA and RNA interference pathway could be used to silence and prevent the mobilization of HIV retroviruses to curb its spread.
rasiRNAs can be used in the laboratory in similar ways as siRNAs to specifically target genomic repeat elements, and used to better study the biology of repeated elements like retrotransposons.
rasiRNAs are also known as repeat-associated small interfering RNAs. They are one of the classes of small noncoding regulatory RNAs that are involved in sequence-specific gene silencing. Other types of small RNAs described to date include small interfering RNAs (siRNAs) and microRNAs (miRNAs). rasiRNAs are 24-27 nucleotides in length and are longer than miRNAs and siRNAs which are typically 20-25 nucleotides long.
rasiRNAs are classified as a subspecies of piwi-interacting RNAs (piRNAs), which form RNA-protein complexes through interactions with Piwi subfamily proteins. rasiRNAs were termed as such because the sequences of these RNAs corresponded to genomic repeats and transposons when they were first discovered in Drosophila melanogaster. Later, RNAs which shared similar features as rasiRNAs were identified in mammalian testes. However, they included RNAs that are not enriched for sequences from transposons and repeats. Hence, this class of RNAs were termed piRNAs and rasiRNAs are considered a specific subtype of piRNAs. (Hartig, 2007)
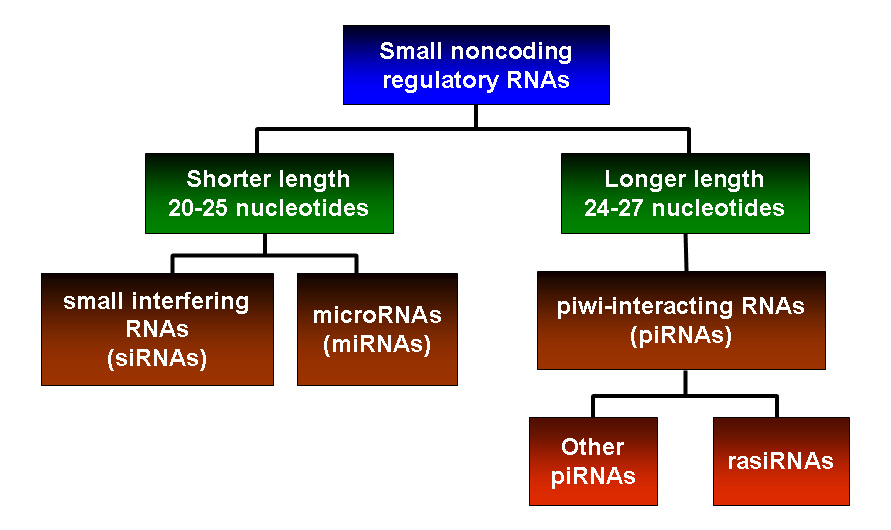
Figure 1. Classification of different classes of small RNAs. rasiRNAs are considered a specific subspecies of piRNAs.
RNA interference regulates heterochromatin and activity of transposable elements
RNAi-like mechanisms were first described in 2000, where scientists noted that transcriptional gene silencing (TGS) and post-transcriptional gene silencing (PTGS) in plants operated via these mechanisms to keep heterochromatic regions condensed and transcriptionally suppressed. It was also believed that one of the natural functions of RNA silencing was to restrict transposon transcription and mobilization, due to evidence that mutations in C.elegans, Drosophila and Chlamydomonas that impair RNA silencing of transgenes or host regulatory genes resulted in elevated activity of transposable elements (Okamoto, 2001)
First hints of rasiRNAs
rasiRNAs were first described in 2001-2002. In Trypanosoma brucei, Djikeng et al. discovered abundant siRNA-like molecules which share sequence homology with 2 retrotransposons that are present in the trypanosome genome INGI and SLACS, both elements having the hallmarks of non-LTR retrotransposons. These siRNA-like small RNAs were about 24-26 nucleotide (nt) long and were present in both sense and antisense polarity. The authors found that these molecules are constitutively expressed and thus speculated that they serve as a housekeeping mechanism to silence retrotransposon transcription and mobilization via the RNAi pathway (Djikeng, 2001).
In Nicotiana and Arabidopsis thaliana plants, Hamilton et al. detected endogenous plant siRNA species corresponding to 3 different retroelements, namely Tnt1 and tobacco TS SINE elements (Nicotiana) and AtSN1 (Arabidopsis). In each case, both the sense and antisense polarities were present and the RNAs were of the larger 24-26 nt class of siRNA, not the smaller 21-23 nt siRNA class. The authors also demonstrate the association of these long siRNA species with DNA methylation and systemic silencing. This provides initial evidence that these siRNAs could serve as systemic signals of RNA silencing and/or mediate DNA methylation. This is consistent with the idea that retrotransposon RNA in plants can be present in a double-stranded siRNA form and processed to lead to the methylation of the corresponding DNA, thereby transcriptionally silencing the retroelement. (Hamilton, 2002)
Coining of the term ‘rasiRNA’ in Drosophila melanogaster
In 2003, Aravin et al conducted small RNA profiling of the different Drosophila melanogaster developmental stages and reported that similar to plants, 2 distinct classes of small RNAs were produced from dsRNA precursors, the shorter 21-23 nt RNAs and the longer 24-26nt RNAs. As the majority of these long RNAs corresponded to the Drosophila genomic repeats or transposons, the term repeat-associated small interfering RNAs (rasiRNAs) was officially coined for this class of RNAs (Aravin, 2003). rasiRNAs were later described in the zebrafish in 2005, where they were reported to associate with Piwi protein homolog Ziwi. (Houwing, 2007)
Distinct biogenesis pathway from other small noncoding RNAs
It was initially believed that rasiRNAs are derived from double-stranded precursors and processed by ribonuclease type III Dicer, similar to miRNA and siRNAs. However, it was later discovered in 2006 that rasiRNAs have a distinct and separate mechanism of biogenesis from miRNAs and siRNAs, in which rasiRNAs likely arise from the antisense strand of repeat elements as a single-stranded precursor, are produced in a Dicer-independent manner and are bound by the Piwi subfamily of Argonaute proteins. Further, scientists proposed a ‘ping-pong’ model for rasiRNA 5’ processing where existing rasiRNA serve to guide the piwi protein complex to define and cleave the 5’ end of the target RNA to form the new rasiRNA. The mechanism for defining the 3’ end is still unknown, but Drosophila studies have suggested that 2 nucleases zucchini and squash are implicated in the process. It was also discovered in 2006 that the 3’ end of rasiRNAs are 2’-O-methylated and this may be possibly a mark defining the 3’ end for Slicer cleavage (see below).
To date, the biogenesis and mechanism of rasiRNAs are still not clearly understood, and this class of RNAs have not been implicated in human diseases. However, it is known that they influence epigenetic modifications of DNA and histones to lead to gene silencing and heterochromatin maintenance. It is very possible that development of some cancers, clinical disorders and developmental failure is due to the dysregulation of the rasiRNA mechanism, leading to abnormal gene expression and epigenetic mechanisms.
Also, retrotransposons move around our genomes and can sometimes result human diseases. It could be that a fraction of diseases may be cause by the deregulation of the rasiRNA pathway.

